A balanced paternal interchromosomal reciprocal insertion between 5q12.1q13.2 and 20p12.3p12.1 resulting in separate genetic conditions in two siblings
TO THE EDITOR
Balanced insertional translocations, where an interstitial segment from one chromosome is inserted into a segment of another chromosome, are rare. Reciprocal insertional translocations are extremely rare, although there are at least 11 other documented cases [Wang et al., 1994; Van Esch et al., 1999; Van Hemel and Eussen, 2000; Bernardini et al., 2008; Ou et al., 2008; Kang et al., 2010; Manolakos et al., 2011; Neill et al., 2011; Harbuz et al., 2013]. We present two siblings with opposing pathogenic copy number variants (CNV) due to a derivative chromosome they inherited from their phenotypically normal father who has two derivative chromosomes due to a balanced reciprocal interstitial exchange involving 5q12.1q13.2 and 20p12.3p12.1. This is a four-break rearrangement with an exchange of interstitial segments from two chromosomes. The first sibling, patient A, with the der(20), had a 20p12.3p12.1 deletion and a 5q12.1q13.2 duplication, consistent with a diagnosis of Alagille syndrome (ALGS) and developmental delay. The second sibling, patient B, with the der(5), had short stature, developmental delay and facial dysmorphism consistent with a 5q12.1q13.2 deletion and 20p12.3p12.1 duplication.
Patient A was born via a spontaneous vaginal delivery at term weighing 2,600 g (<10th centile). Investigations performed soon after birth revealed an elevated direct bilirubin of 45 umol/L and gamma glutamyl-transferase (GGT) of 1551 U/L with a normal ultrasound scan of the biliary tree and liver. At 3 years and 6 months of age her weight was 13 kg (10th centile) her height was 91 cm (5th centile) and her head circumference was 49 cm (50th centile). She had global development delay and bilateral branch pulmonary arterial stenosis was detected on a surveillance echocardiogram. An ophthalmic review and spine X-ray were normal. On examination, she had long fingers with single palmar creases. Dysmorphic facial features included a small mouth, a small pointed chin, squared nasal tip, high and broad nasal root, up-slanting palpebral fissures, medially sparse eyebrows, and gum hypertrophy. Her ears were simple and prominent with unattached ear lobes. Patient B is the younger sister of patient A and was born at 36 weeks gestation after a pregnancy complicated by severe idiopathic polyhydramnios. She developed seizures at 11 months of age and a brain MRI demonstrated left-sided periventricular heterotopia. She had leg length discrepancy due to a dysplastic left acetabulum and hypoplastic left femoral epiphysis. She had developmental delay and at 3 years of age her head circumference was 49 cm (50th centile) and her height was 80 cm (<1st centile). On examination, she had up-slanting palpebral fissures with a unilateral epicanthic fold and a broad nasal bridge.
FISH studies for patient A demonstrated a heterozygous deletion (ish del[20][p12.2p12.2][DJ1099D15-]), consistent with a diagnosis ALGS. The 20p deletion was confirmed on an Agilent CGH 60 K and 180 K microarray, which also documented a 5q interstitial duplication: ISCN 2013 arr [hg19] 5q12.1q13.2(62,855,643–71,728,887)×3 approximately 8.8 Mbp, 20p12.3p12.1(8,058,870–15,317,311)×1 approximately 7.2 Mbp. FISH analysis for patient B revealed three copies of the DJ1099D15 probe; two on chromosome 20 and one on chromosome 5. The 20p duplication was confirmed on an Agilent CGH 60 K and 180 K microarray, which also documented a 5q interstitial deletion: ISCN 2013 arr [hg19] 5q12.1q13.2(62,839,696–71,673,176)×1 approximately 8.8 Mbp, 20p12.3p12.1(8,058,870–15,240,340)×3 approximately 7.2 Mbp. No abnormalities were detected on an Agilent CGH 60 K microarray or FISH studies for the children's mother. For the father, an Agilent CGH 60 K microarray gave a normal result. FISH analysis revealed two copies of the DJ1099D15 probe; one on chromosome 5 and one on chromosome 20. The similar lengths of the two exchanged chromosome segments complicated standard cytogenetic studies. Whole chromosome paint (WCP) studies demonstrated the presence of material from chromosome 20 at 5q and the presence of material from chromosome 5 at 20p. Figure 1 shows the paternal karyotype and WCP data (other FISH data not shown). Thus, the father carries two derivative chromosomes due to a balanced reciprocal interstitial exchange, involving segments 5q12.1q13.2 and 20p12.3p12.1. This paternal karyotype is supported by FISH observations as follows: ish der(5)ins(5;20)(wcp5+,wcp20+,DJ1099D15 + ), der(20)ins(20;5)(wcp5+,wcp20+,DJ1099D15−). Ideograms and formal karyotypes for the rearrangement seen in the father and both children are depicted in Figure 2.

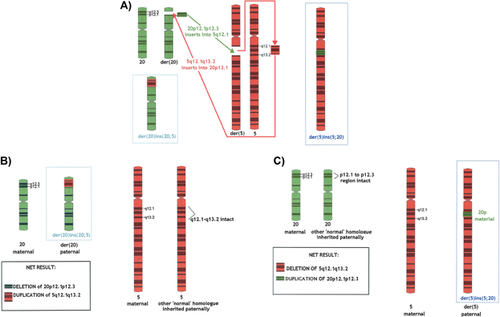
The deleted 20p12.3p12.1 region found in patient A contains 18 genes, 7 of which have been associated with disease-causing mutations [OMIM]: PLCB1 (MIM 607120), PLCB4 (600810), SNAP25 (600322), MKKS (604896), JAG1 (601920), NDUFAF5 (612360), and FLRT3 (604808). Note however, that the MIM listings include many recessive loci, unlikely to be relevant in the context of heterozygous deletion (and duplication). Of the genes listed above, MKKS and NDUFAF5 are not regarded as likely haploinsufficient (HI) within the DECIPHER database [DECIPHER]. Haploinsufficiency of JAG1 is found in approximately 5% of patients with ALGS [Warthen et al., 2006], and accordingly, DECIPHER ranks JAG1 among the ∼1% of genes most likely to be HI. Heterozygous mutation in PLCB4 has been associated with auriculocondylar syndrome (ACS), or “question-mark ear syndrome” although haploinsufficiency of PLCB4 does not appear to be causative [Gordon et al., 2013] and Patient A lacked clinical features of ACS. Of course, Patient A also has duplication of 5q12.1q13.2, and this imbalance (via any “triplosensitive” genes) was also likely to contribute to her phenotype. Given the duplication's size and number of genes included: it was classified on initial microarray testing as “likely clinically significant.” However, since Patient A presented predominantly with features of ALGS including liver dysfunction, branch pulmonary stenosis, and dysmorphic facial features, the effects of the duplication may be relatively slight. This is consistent with the long-held view in human cytogenetics that dosage increases (i.e., partial trisomies) have lesser phenotypic effects than partial monosomies of equivalent size [Daniel, 1979]. We found no evidence for triplosensitivity, specific to any particular gene within the 5q12.1q13.2 region; although there is minimal gene-specific data on the publically available databases at present [ClinGen].
The deleted 5q12.1q13.2 region found in Patient B contains 41 genes, 8 of which have been associated with disease-causing mutations [OMIM]: HTR1A (MIM 109760), PIK3R1 (171833), MARVELD2 (610572), OCLN (602876), SMN2 (601627), SMN1 (600354), MCCC2 (609014), and CARTPT (602606). Jaillard et al. [2011] reported four patients with similar chromosomal deletions encompassing a shared 2.6 Mbp region at 5q12.1 (59,354,365–61,985,998) with common clinical features including developmental delay, coarse facies and ocular anomalies. Patient B's deletion did not include this 2.6 Mbp region at 5q12.1; however, Patient 1 reported by Jaillard et al. [2011] did have a similar deletion to Patient B and they shared phenotypic similarities including unexplained polyhydramnios, seizures, and growth retardation. The overlapping deleted region between Patient 1 and Patient B was located at 5q12.1q13.2 and was 6 Mbp in size (62,839,696–68,864,273). Cetin et al. [2013] reported a patient who had seizures, developmental delay, a normal brain MRI and short stature with a deletion at 5q12.1q12.3 that included only HTR1A and RNF180 (62,850,767–63,738,454). Cetin et al. [2013] concluded that RNF180 deletion might be associated with mental and motor retardation and that HTR1A deletion might be associated with seizures. Holder and Cheung [2015] recently refined a region likely to be causative of growth restriction to an approximately 800 Kbp section at 15q12.3 that was also encompassed by our patient (64,520,143–65,309,488). It was therefore felt the phenotype of our patient was predominantly explained by the presence of the 5q12.1q13.2 deletion. The duplication at 20p12.3p12.1 was also likely to contribute to her phenotype and was classified as “likely clinically significant” on the initial microarray due to the size and number of genes involved. Relevant databases offer no evidence for triplosensitivity, attributable to any specific gene within the 20p12.3p12.1 region [ClinGen], although up-slanting palpebral fissures have been reported in other patients with a similar 20p duplication [de Ravel et al., 2003].
Chromosomal microarray studies in both parents were normal and FISH studies were required to establish the father's complex but balanced cytogenetic rearrangement. The birth prevalence of microscopically visible insertions has been estimated at approximately one in 80,000 [Van Hemel and Eussen, 2000]. However, their detection has increased with the advent of microarray technology, and newer estimates suggest a frequency of approximately one in 500 [Manolakos et al., 2011; Harbuz et al., 2013]. Balanced parental insertional translocations detectable by FISH underlie approximately 2.1% of apparently de novo interstitial CNVs [Nowakowska et al., 2012]. When the propositus has an insertional translocation, 10% may be due to the inheritance of a derivative chromosome from a parent with a balanced rearrangement [Harbuz et al., 2013]. In studies of such insertions, follow-up parental studies are not always available, or complete, and so this 10% estimate may be regarded as a minimum. This case highlights the recommendation that, where possible, FISH analysis is a necessary adjunct to microarray-based cytogenetics, to fully exclude a balanced chromosomal rearrangement in the parents of a child with an apparently de novo interstitial deletion and duplication of pathological interest that is not due to a segmental mediated rearrangement.
WEB RESOURCES
ClinGen Dosage Sensitivity Map; ClinGen, http://www.ncbi.nlm.nih.gov/projects/dbvar/clingen/
Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources; DECIPHER, http://decipher.sanger.ac.uk
Online Mendelian Inheritance in Man; OMIM, http://www.ncbi.nlm.nih.gov/omim