A novel TUBB3 mutation in a sporadic patient with asymmetric cortical dysplasia
Abstract
Recent advances in molecular technology have led to the discovery of several genes related to human malformations of cortical development (MCDs). The beta-tubulin class III gene (TUBB3) was identified as a gene responsible for MCDs. Although mouse-model experiments have not revealed any findings of neuronal migration disorders, human TUBB3 mutations have been identified in patients with congenital fibrosis of the extraocular muscles. Since the discovery of a TUBB3 mutation, only 15 mutations have been identified. In this study, comprehensive mutation screening through next-generation sequencing identified a novel TUBB3 mutation (p.Ser230Leu) in a sporadic patient with moderate developmental delay associated with mild MCD. Compared to patients with the alpha-tubulin class 1a gene (TUBA1A) mutations, patients with TUBB3 mutations show milder phenotypic manifestations and milder MCD. Therefore, patients with milder MCD manifestations may be under-diagnosed, and TUBB3 mutations may be rarely identified. Additional genotype–phenotype information should be accumulated for further understanding of the TUBB3 functional relevance. © 2016 Wiley Periodicals, Inc.
INTRODUCTION
Human brain structure is complex compared to that of rodents. During the development of the human brain, many genes contribute to neuronal migrations. Disruptions in normal brain development lead to malformations of cortical development (MCDs). The most frequently observed genetic alteration in patients with MCD is a chromosomal deletion of 17p13.3, where the platelet-activating factor acetylhydrolase 1b regulatory subunit 1 gene (PAFAH1B1, also known as LIS1) is located [Shimojima et al., 2015, 2010]. Other than LIS1, many additional genes contribute to MCD. Recent advances in molecular technology have led to the discovery of several genes responsible for MCD. In particular, an increasing number of MCDs are associated with mutations in genes of the tubulin family, including the alpha-tubulin class 1a gene (TUBA1A), the beta-tubulin 2B class IIb gene (TUBB2B), and the beta-tubulin class III gene (TUBB3) [Tischfield and Engle, 2010; Tischfield et al., 2011; Romaniello et al., 2015]. Recently, disorders caused by mutations in the tubulin gene family were recognized as tubulin-related disorders or tubulinopathies [Poirier et al., 2010; Tischfield et al., 2010; Bahi-Buisson et al., 2014]. Among these, diseases caused by mutations in TUBB3 are rare, and only 15 TUBB3 mutations have been identified [Demer et al., 2010; Chew et al., 2013; Fallet-Bianco et al., 2014; Hong et al., 2015].
Here, we report a novel TUBB3 mutation in a sporadic patient with severe developmental delay associated with unilateral cortical dysplasia.
Case Report
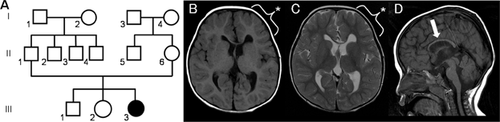
A girl, aged 1 year and 4 months, was born at 40 gestational weeks with a birth weight of 2,594 g (25–50th percentile), length of 49 cm (50–75th percentile), and occipitofrontal circumference (OFC) of 31 cm (<10th percentile), indicating microcephaly. She was the third child of healthy parents, and her older brother and older sister were healthy (Fig. 1A). She had shown developmental delay since early infancy: she held up her head at 4 months old and rolled over at 8 months. Conventional G-banding showed a normal female karyotype of 46,XX.
At present, the patient shows growth failure with a height of 74.4 cm (10–25th percentile), weight of 7.4 kg (<3rd percentile), and OFC of 43 cm (<3rd percentile). She is microcephalic with flattened occiput. Distinctive facial findings of downward slanting palpebral fissures, epicanthal folds, low-set ears, high-arched palate, and micrognathia are noted. Her hands and feet are small. She can sit by herself but cannot crawl by herself. She shows visual pursuit and can communicate with gestures. She can masticate and swallow smoothly. However, she cannot speak any meaningful words. From these findings, we have determined that she shows severe psychomotor developmental delay.
Brain magnetic resonance imaging (MRI) analysis at the age of 2 years and 5 months showed asymmetric dilatation of the lateral ventricles and volume loss of the corpus callosum (Fig. 1B–D). Left-frontal cortical dysplasia was later confirmed by retrospective and careful re-evaluation.
Molecular Analysis
For a genetic diagnosis, blood samples were obtained from the patient and her parents after receiving informed consent. This study was performed in accordance with the Declaration of Helsinki, and the ethics committee of Tokyo Women's Medical University approved this study. Genomic DNA was extracted from blood samples and used for further examination. Genomic copy number aberrations were examined using an Agilent SurePrint G3 Human CGH Microarray Kit 8 × 60 K (Agilent Technologies, Santa Clara, CA), as described previously [Shimojima et al., 2010; Shimojima et al., 2015] and demonstrated no pathogenic aberrations. Subsequently, targeted resequencing was performed using TruSight One v1.0 Sequencing Panel (Illumina, San Diego, CA), as described previously [Yamamoto and Shimojima, 2015]. Briefly, this panel includes 125,396 probes aimed to capture 11,946,514 bp of targeted exon regions consisting of 4,813 genes associated with known clinical phenotypes. After constructing the sequence library using 50 ng genomic DNA, the MiSeq next-generation sequencer (Illumina) was used to sequence 151-bp paired-end reads, according to the manufacturer's instructions. The extracted data were mapped to a reference genome (GRCh37/hg19) by using BWA Enrichment v1.0 cloud software (Illumina). Total aligned bases consisted of 3.1 Gb of sequence, and a mean coverage depth of 123.8× was obtained (the target coverage at 20× was 96.1%). In total, 8,228 variants were extracted, annotated, and filtered using the Variant Studio software (Illumina).
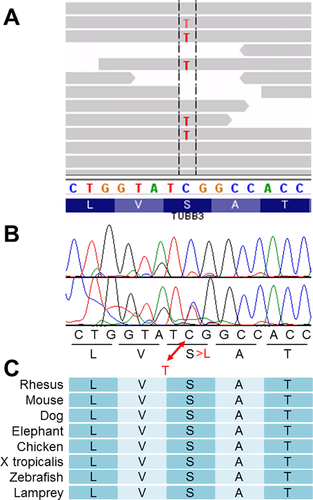
We excluded variants associated with the following conditions from our analysis: variants localized to non-conserved regions, variants listed in dbSNP database and/or JSNP database, variants with population frequencies >1%, variants with altered allele frequencies <33%, and variants corresponding to synonymous substitutions. Subsequently, variants listed by SIFT as “deleterious” and PolyPhen-2 as “damaging” were selected. This left nine missense variants within nine genes (Supplemental Table SI). We analyzed the trio samples (including parental samples) by Sanger sequencing and confirmed that only the variant in TUBB3, NM_006086.3(TUBB3):c.689C>T [NP_006077.2(TUBB3):p.Ser230Leu] (Fig. 2B), was of de novo origin. The functional relevance of the TUBB3 variant was evaluated and this was considered as a possible candidate and the others were eliminated due to inheritance from either of parents. The TUBB3 alteration was observed in 45.9% of the reads (altered depth/read depth = 117/255), indicating no germline mosaicism (Fig. 2A). We checked three dimensional conformation of the TUBB3 protein by web-based software SWISS-MODEL (http://swissmodel.expasy.org/). Although the affected codon is in the alpha helix domain conserved among species (Fig. 2C), the altered codon did not change three dimensional conformation of TUBB3 (Supplemental Fig. S1). The prediction scores for this variant were evaluated by SIFT as 0.01 (deleterious) and by PolyPhen-2 as 0.936 (probably damaging).
DISCUSSION
In this study, we successfully identified a de novo TUBB3 mutation in a patient with severe developmental delay associated with MCD. The patient's clinical and radiological findings were retrospectively evaluated after identification of TUBB3 mutation. The patient showed severe developmental delay but did not show pyramidal signs. This is reasonable because the affected cortical region was restricted to the frontal region (Fig. 1B, C). Such asymmetric radiological findings were compatible with those previously reported in patients with TUBB3 mutations [Poirier et al., 2010].
In 2010, TUBB3 was identified as a gene responsible for MCD [Poirier et al., 2010]. Although mouse-model experiments have not revealed any neuronal migration disorders, human TUBB3 mutations have been identified in patients with congenital fibrosis of the extraocular muscles type 3 (CFEOM3) or polyneuropathy/developmental delay [Tischfield et al., 2010]. Since the first discovery of a TUBB3 mutation in patients with neurological impairments, only 15 mutations have been identified as mentioned above. The mutation, p.Ser230Leu, identified in this study has never reported previously. The present mutation and the 15 previously reported mutations can be seen on the primary structure of TUBB3 (Fig. 3). TUBB3 mutations were observed in patients with CFEOM3 or MCD. Regardless of the phenotypic features, reported mutations are distributed across the entirety of TUBB3, and there is no clear genotype–phenotype correlation.
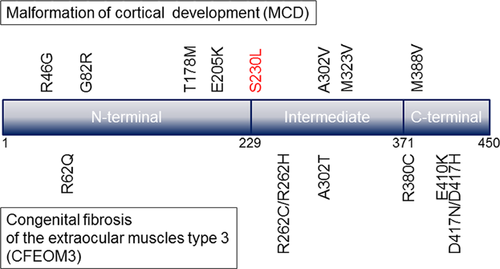
Compared to TUBA1A, the number of reported mutations in TUBB3 is extremely small [Bahi-Buisson et al., 2014]. Because the two genes are almost the same in length, gene size does not appear to cause the scarcity of TUBB3 mutations. Rather, the rarity of TUBB3 mutations may be due to the milder phenotypic appearance. Patients with TUBA1A mutations generally show more severe clinical features (most patients are bedridden) and more severe MCDs, such as lissencephaly [Okumura et al., 2013; Shimojima et al., 2014]. Comparatively, patients with TUBB3 mutations show milder phenotypic manifestations and milder MCDs, such as brain asymmetry, partial cortical dysgenesis, and malformations in basal ganglia. Therefore, patients with milder MCD manifestations may be under-diagnosed, and TUBB3 mutations may be rarely identified. Additional genotype–phenotype information should be accumulated for further understanding of the TUBB3 functional relevance.
ACKNOWLEDGMENTS
We would like to express our gratitude to the patient and her family for their cooperation. This work was mainly supported by a grant from the Precursory Research for Embryonic Science and Technology (PRESTO) program of the Japan Science and Technology Agency (JST), Kawaguchi, Japan. It was partially supported by a Grant-in-Aid for Young Scientists (B) (24791090) from the Japan Society for the Promotion of Science (JSPS), a grant from the Japan Epilepsy Research Foundation (JERF), and a grant from Kanae Foundation for the promotion of Medical Science in Japan (K.S.). In addition, it was partially supported by the Practical Research Project for Rare/Intractable Diseases from Japan Agency for Medical Research and Development (AMED) and JSPS KAKENHI Grant Number 15K09631.