A homozygous HOXD13 missense mutation causes a severe form of synpolydactyly with metacarpal to carpal transformation
Abstract
Synpolydactyly (SPD) is a rare congenital limb disorder characterized by syndactyly between the third and fourth fingers and an additional digit in the syndactylous web. In most cases SPD is caused by heterozygous mutations in HOXD13 resulting in the expansion of a N-terminal polyalanine tract. If homozygous, the mutation results in severe shortening of all metacarpals and phalanges with a morphological transformation of metacarpals to carpals. Here, we describe a novel homozygous missense mutation in a family with unaffected consanguineous parents and severe brachydactyly and metacarpal-to-carpal transformation in the affected child. We performed whole exome sequencing on the index patient, followed by Sanger sequencing of parents and patient to investigate cosegregation. The DNA-binding ability of the mutant protein was tested with electrophoretic mobility shift assays. We demonstrate that the c.938C>G (p.313T>R) mutation in the DNA-binding domain of HOXD13 prevents binding to DNA in vitro. Our results show to our knowledge for the first time that a missense mutation in HOXD13 underlies severe brachydactyly with metacarpal-to-carpal transformation. The mutation is non-penetrant in heterozygous carriers. In conjunction with the literature we propose the possibility that the metacarpal-to-carpal transformation results from a homozygous loss of functional HOXD13 protein in humans in combination with an accumulation of non-functional HOXD13 that might be able to interact with other transcription factors in the developing limb. © 2015 Wiley Periodicals, Inc.
INTRODUCTION
Mutations HOXD13 cause synpolydactyly (SPD1, OMIM 186000), a dominantly inherited limb malformation characterized by cutaneous webbing between the third and fourth digit, variably associated with postaxial polydactyly in the same digits. The phenotype can vary in penetrance and expressivity within families and even within the limbs of a patient. At the feet, HOXD13 mutations are often associated with syndactyly of the fourth and fifth toe and postaxial polydactyly [Goodman et al., 1998]. HOXD13 is a highly conserved transcription factor that belongs to the homeotic genes of the HOXD cluster, which pattern the vertebrate body along the anterior-posterior axis and the limb along the proximo-distal axis. HOXD13 (NM_000523.3) codes for a 343 amino acid protein that carries a 15 amino acid polyalanine repeat in the N-terminal part and the DNA-binding homeodomain at its C-terminus (amino acids 276–335) [Brison et al., 2014].
Mutations in HOXD13 can be classified in three molecular groups: putative null or loss-of-function (LOF) mutations (e.g., nonsense or frameshift), expansions of the N-terminal polyalanine repeat, and missense mutations. Whereas the first two groups of mutations are exclusively associated with SPD phenotypes, missense mutations in HOXD13 can cause SPD, various brachydactylies and other, unusual limb malformations [Johnson et al., 2003; Ibrahim et al., 2013; Brison et al., 2014].
Nonsense and frameshift mutations are thought to cause haploinsufficinency of the HOXD13 protein due to nonsense-mediated decay, although no experimental validation for this has been reported. These LOF-associated SPDs have a variable phenotype with incomplete penetrance in heterozygous patients but are fully penetrant in homozygous individuals [Goodman et al., 1998; Kan et al., 2003; Kurban et al., 2011].
The most common type of HOXD13 mutations are expansions of the 15-residue N-terminal alanine repeat [Muragaki et al., 1996]. Expansions of +7 to +14 alanines have been reported in over 40 families with varying penetrance and expressivity of the SPD phenotype [Malik and Grzeschik, 2008], which increases in severity with the length of the alanine expansion [Goodman et al., 1997]. It has been shown that the polyalanine-tract expansions lead to cytoplasmatic retention and aggregation of mutant HOXD13 protein. Furthermore, in vitro studies demonstrated that the mutant protein acts in a dominant-negative manner, as it also leads to cytoplasmatic aggregates, which were shown to contain not only wild-type HOXD13 proteins, but also other polyalanine containing proteins such as HOXA13 or RUNX2 [Albrecht et al., 2004].
Finally, thirteen missense mutations in HOXD13 have been reported, six of which reside in the DNA-binding homeodomain [Brison et al., 2014]. Whereas missense mutations outside of the homeodomain are associated with SPD phenotypes, missense mutations inside the homeodomain can cause multiple phenotypes that include SPD-like dysplasias, and other phenotypes, such as Brachydactylies type D and E, syndactyly type V and an brachtydactyly–oligodactyly phenotype [Caronia et al., 2003; Johnson et al., 2003; Zhao et al., 2007; Ibrahim et al., 2013]. These non-SPD phenotypes result from gain-of-function effects that alter the characteristics of the HOXD13 protein [Ibrahim et al., 2013].
The majority of patients carrying HOXD13 mutations are heterozygous and usually follow an autosomal-dominant inheritance pattern with incomplete penetrance. Patients homozygous for HOXD13 mutations have been described in four families with polyalanine expansions and one with a nonsense mutation [Akarsu et al., 1995; Muragaki et al., 1996; Horsnell et al., 2006; Kurban et al., 2011]. In individuals homozygous for the polyalanine expansions the SPD phenotpye is fully penetrant. Affected individuals have very short metacarpals and phalanges. The metacarpals loose the typical tubular structure and exhibit a rounded morphology reminiscent of carpal bones [Kuss et al., 2014, 2009]. This phenotype is recapitulated in a mouse model that carries a +7 alanine expansion [Johnson et al., 1998]. As in humans, the homozygous mice show a severe poyldactyly and very short bones with a metacarpal-to-carpal transformation. In contrast, heterozygous mice do not show any phenotype. The only other patients homozygous for a HOXD13 mutation are from a large Pakistani family with a p.Q248X nonsense mutation that truncates the protein and removes the DNA-binding domain [Kurban et al., 2011]. Although heterozygous individuals have a typical SPD-phenotype with incomplete penetrance, homozygous patients from this family display a phenotype that seems to cause a metacarpal-to-carpal transformation, raising the possibility that the phenotype results primarily from the loss of functional HOXD13 protein.
Here, we describe a novel missense mutation in the DNA-binding homeodomain of HOXD13 causing a severe form of SPD with metacarpal-to-carpal transformation. This is to our knowledge the first report of a homozygous missense mutation in HOXD13 that shows no obvious phenotype in heterozygous individuals, but causes a severe metacarpal-to-carpal transformation in the homozygous index patient. Following in silico analysis of the mutation, we show that the mutant HOXD13 protein loses its ability to bind the HOXD13 DNA in vitro.
Therefore we propose that in humans the metacarpal-to-carpal transformation phenotype is caused by homozygous loss of functional HOXD13 protein in combination with an accumulation of non-functional HOXD13, which might be able to interact with other transcription factors in the developing limb.
METHODS
Human Material
Venous blood samples were obtained from the patients by standard procedures. Written informed consent was obtained from all individuals studied to participate in this study. This study was approved by the Charité Universitätsmedizin Berlin Ethics Committee.
Patients
In this study, an Iranian family with a severe synpolydactyly phenotype and no family history of limb malformations was investigated. Preoperative photographs of the pro-band at the age of 2, as well as post-operative photo- and radiographs at the age of 4 years were taken. Additionally, digital photographs of his parents were taken as control to evaluate the phenotype.
Next Generation Sequencing and Mutation Detection
Since no clinical diagnosis could be established the patient was included in a next generation sequencing (NGS) exome study. Genomic DNA was isolated from peripheral blood samples and, following Agilent SureSelect All Exon kit V4 enrichment, subjected to next generation sequencing (paired end 150 bp). Subsequently, a computational method termed Phenotypic Interpretation of eXomes (PhenIX), which evaluates variants based on population frequency and predicted pathogenicity and then ranks the genes according to variant score and a clinical relevance score, was used to prioritize candidate genes [Zemojtel et al., 2014]. To calculate the clinical relevance score we entered the following human phenotype ontology [Robinson et al., 2008] terms HP0010708 and HP0010713.
Sanger Sequencing
Genomic DNA from the patient and his parents was extracted from peripheral blood leukocytes using standard protocols. Coding region and flanking intronic sequences of the exons of HOXD13 (NM_000523.3) were amplified and sequenced using standard protocols. Primers and PCR conditions are available upon request. The sequencing results were processed with the 4Peaks software.
Electrophoretic Mobility Shift Assays
Cloning and expression of wild-type and mutant homeodomains in E. coli were performed as described previously. [Ibrahim et al., 2013] EMSAs were performed using 20 μl binding buffer (100 mM NaCl, 2 mM MgCl2, 0.1 mg/ml BSA, 4 mM spermidine, 25 mM HEPES, pH 7.5, Roche complete protease inhibitor), and 1.5 μl poly(dIdC) (100 ng/μl) as well with varying amounts of purified homeodomain (3, 6, 9, 12, 15, 20 ng) were added. Reactions were incubated on ice for 5 min, then 1 μl of the Cy3-labeled double-stranded oligonucleotide (5′CY3-ggatcCCAATAAAAtcggc-3′) (0.75 pmol/μl) followed by 15 min incubation at room temperature (20°C). For testing sequence specificity, 15 pmol (15 pmol/μl) unlabelled competitor oligo (wild-type or mutant 5′-ggatcCCcAgcAcAtcgg -3′) were added to the reaction. Before loading of the sample onto the gel, 2 μl loading buffer (40% glycerol + 0.01% bromphenol blue) were added. Electrophoresis was performed on an 8% native polyacrylamide gel in 1× TBE and scanned on a FLA-5000 Scanner (Fuji).
Mutation Modeling
The NMR structure of the paralog HOXA13 bound to a DNA helix (PDB ID 2LD5, [Structural basis for sequence specific DNA binding and protein dimerization of HOXA13. http://www.rcsb.org/pdb/explore.do?structureId=2ld5]) was used to depict the threonine to arginine mutation. In the paralog HOXA13 Thr44 is equivalent to the Thr38 in HOXD13. The in silico mutagenesis was modelled with PyMol v1.3.
RESULTS
Clinical Report
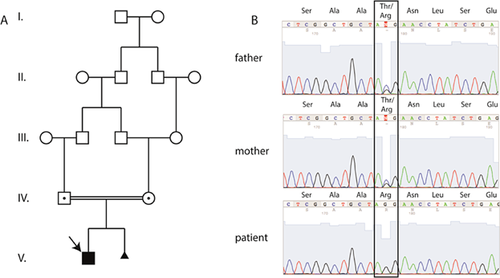
The index patient is a 4-year old boy with severe synpolydactyly involving toes and fingers two to five of both extremities (Fig. 1A). After an uneventful pregnancy, limb deformities were first noted at birth. Four operations were performed on the patient's hands and feet (Fig. 1A), and a cryptorchidism was also operated. Anthropometric measurements at the age of four revealed normal stature, weight and head circumference. His parents are healthy first cousins (Fig. 1C, D) and encountered the same limb phenotype in the 2nd pregnancy, diagnosed via 3D ultrasonography, upon which the pregnancy was terminated. There is no history of limb malformations in the family (Fig. 2A).
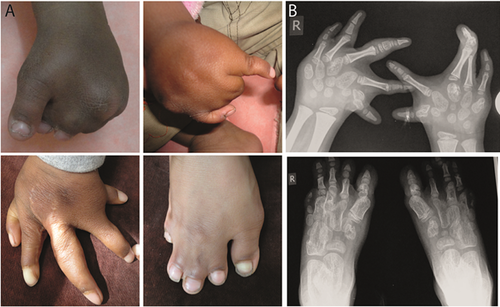
Post-operative radiographic studies at the age of four years confirmed the patient to be affected by a severe form of SPD, with a post-operatively remaining bony fusion between the 3rd and 4th digits of the right hand (Fig. 1B). Despite a fully cutaneous fusion of the second to fifth toes, the radiographs did not reveal any bony fusions.
Additionally, the radiographs revealed an abnormal morphology of all metacarpals and metatarsals (Fig. 1B). Both, metacarpals and—tarsals do not show the normal tubular shape and neither epiphysis nor medullary cavity can be seen on x-rays. The bones rather appear to take on a rounded shape reminiscent of carpal bones. A similar metacarpal-to-carpal transformation phenotype was described by Lorini et al. in cases with homozygous polyalanine-expansion mutations [Villavicencio-Lorini et al., 2010].
Mutation Detection
The patient was included in a whole exome sequencing study. Following variant calling, PheniX analysis (search terms: HP 0010708; HP0010713) of the variants ranked a homozygous point mutation (c.938C>G) in HOXD13 as top hit in the list of disease causing mutations (Table I). This mutation leads to a threonine to arginine substitution at position 313 (p.T313R) of the HOXD13 protein, corresponding to position 38 of the DNA-binding homeodomain. We will therefore refer to this mutation as p.T38R.
| Gene | Gene relevance score | Variant score | Total score | |
|---|---|---|---|---|
| 1 | HOXD13 | 1.000 | 1.000 | 1.000 |
| 2 | LRBA | 0.504 | 1.000 | 0.752 |
| 3 | HLA-DRB1 | 0.423 | 0.951 | 0.687 |
| 4 | LRP2 | 0.210 | 1.000 | 0.605 |
| 5 | ADAR | 0.210 | 0.960 | 0.585 |
| 6 | SEC63 | 0.105 | 0.900 | 0.503 |
| 7 | MCCC1 | 0.004 | 1.000 | 0.502 |
| 8 | KCNC3 | 0.002 | 1.000 | 0.501 |
| 9 | CAST | 0.000 | 1.000 | 0.500 |
| 10 | XKRX | 0.000 | 1.000 | 0.500 |
Since both parents were not affected but first cousins, we performed cosegregation analysis via Sanger sequencing and confirmed both parents to be heterozygous for the point mutation (Fig. 2B). In addition, Sanger sequencing confirmed the patient to be homozygous for the missense mutation.
Functional Evaluation of the HOXD13T38R Variant
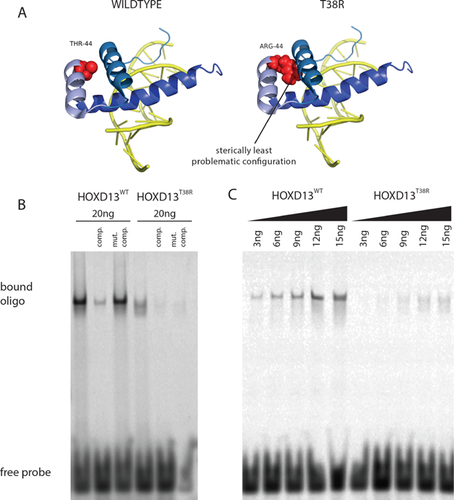
The T38R substitution resides in second helix of the homeodomain pointing inwards. Figure 3A shows a NMR derived structure of a HOXA13 homeodomain (91% identical to HOXD13) bound to DNA (PDB ID: 2LD5) that highlights the mutated threonine. We then introduction the T38R mutation in silico, which is a relatively large and positively charged amino acid. The introduction of this amino acid resulted in severe steric problems. Figure 3A shows the protein configuration with the least steric hindrances. The T38R substitution probably severely impairs the correct folding of the homeodomain and thereby inhibits the ability of the protein to bind DNA effectively, in consequence leading to a loss of DNA-binding ability.
The HOXD13T38R Mutant Is not Able to Bind the HOXD13 Binding Site In Vitro
To determine whether the mutant loses its ability to bind DNA, we performed Electrophoretic Mobility Shift Assays (EMSA) with purified wild-type and T38R mutant homeodomains and dsDNA-oligonucleotides carrying the characteristic HOXD13 binding site. As shown in Figure 3B and C, HOXD13wt binds the probe in a sequence-specific manner producing a sharp, shifted band in the gel that increases in a concentration dependent manner. In contrast, the HOXD13T38R mutation did not produce a sequence-specific gel shift. However, at high protein concentrations a smeary signal appeared instead of a sharp band, indicating that the HOXD13T38R homeodomain is not able to bind the HOXD13 binding site.
DISCUSSION
Non-syndromic Synpolydactyly (SPD) is clinically and genetically heterogenous limb defect. Heterozygous mutations in the HOXD13 gene have been shown to cause SPD with a broad spectrum of inter- and intra-familial phenotypic manifestations [Brison et al., 2014]. So far, four families with individuals carrying homozygous HOXD13 mutations have been reported. Heterozygous individuals typically display an SPD-phenotype with incomplete penetrance, whereas homozygous individuals exclusively show an additional metacarpal-to-carpal transformation phenotype that is fully penetrant. In this study, we identified and analyzed a novel missense mutation in the DNA-binding homeodomain of HOXD13 that causes severe SPD with metacarpal-to-carpal transformation in the homozygous index patient, but is without obvious phenotype in the heterozygous parents.
The c.938C>G transition in exon two of HOXD13 results in a p.T313R substitution, located at position 38 of the DNA-binding homeodomain of HOXD13. The homeodomain is 100% conserved between human and chicken on protein-level, but sequence comparison between several human HOX homeodomains indicates that the amino acid residue 38 is variable between related homeodomains. However, the chemical nature of the amino acids at the corresponding residue 38 are similar. All HOX homeodomains have relatively small, aliphatic amino acids such as valine, leucine, or isoleucine at position 38. The HOX13 paralogs are the only ones harboring a threonine at this position, which is weakly polarized via a hydroxyl-group. As we show in our mutation modelling, the introduction of the large, positively charged arginine is likely not compatible with the correct folding of the homeodomain and would thereby lead to a loss of DNA-binding ability in the mutant protein, as supported by our EMSA results.
To date, seven distinct missense mutations in the HOXD13 homeodomain have been shown cause dominantly inherited limb malformations. Three LOF-alleles, R31G, R31Q, and R31W, cause a SPD phenotype, with incomplete penetrance in the case of R31G and R31W [Debeer et al., 2002; Dai et al., 2014]. The four remaining missense mutations are GOF-alleles and do not cause SPD, but Brachydactyly type E and D (S41C and I47L), Syndactyly Type V (Q50R), and a complex brachydactyly-syndactyly phenotype (Q50K) [Caronia et al., 2003; Johnson et al., 2003; Zhao et al., 2007; Ibrahim et al., 2013].
In contrast to the dominant inheritance of all other HOXD13 mutations, the T38R inheritance appears to be autosomal-recessive. However, dominant inheritance with incomplete penetrance in the parents also has to be considered, similar to the previously reported weak penetrance of the R31G and R31W mutations.
Homozygous HOXD13 Mutations Cause Metacarpal-to-Carpal Transformation
Here we report to our knowledge the first homozygous missense mutation in HOXD13 that is associated with the metacarpal-to-carpal transformation phenotype. To date, it remains unclear whether the metacarpal-to-carpal transformation is caused by the deleterious effect of homozygous polyalanine-mutations on other TFs or rather results from the complete loss of functional HOXD13 protein. Contradictory effects of two mouse models nourished this uncertainty. Whereas a homozygous HOXD13 knockout mouse model reduced digit length, but did not show metacarpal-to-carpal transformation [Dollé et al., 1993], a mouse model for the polyalanine-tract expansions recapitulated the patient phenotype with the same metacarpal-to-carpal transformation in homozygous state [Johnson et al., 1998], However, unlike all human mutations, which are inherited dominantly, both mutants did not produce any heterozygous phenotype.
The difference between mouse model and human patients might be explained by the type of introduced mutation. The Hoxd13 knockout mouse model does not produce any remaining HOXD13 protein. In contrast, the polyalanine-expansions (in mice and men), the human p.Q243X truncation, and the p.T38R mutations possibly result in a mutant HOXD13 protein, that could accumulate in the nucleus and might be able to interact with other transcription factors in the developing limb causing mis-regulation.
Therefore we suggest that the metacarpal-to-carpal transformation might be due to an accumulation of non-functional HOXD13 protein in the nucleus in the absence of functional HOXD13 protein. The non-functional HOXD13 protein might then interact with other transcription factors in the developing limb causing the metacarpal-to-carpal transformation.
ACKNOWLEDGMENTS
We would like to thank the family for their collaboration and contribution to this project. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG) to SM. MS was supported by a fellowship of the Berlin-Brandenburg School for Regenerative Therapies (BSRT), Berlin, Germany. Contributors: Patient recruitment and phenotyping: DMI, NT, MS, AS, SM, and MS. NGS experiments and data analysis DMI, NT, ACS, JH, SM, AK and MS. All the authors contributed in writing and reviewing the manuscript.




