Compound heterozygous PKHD1 variants cause a wide spectrum of ductal plate malformations
Abstract
Ductal plate malformations (DPM) present with a wide phenotypic spectrum comprising Von Meyenburg complexes (VMC), Caroli disease (CD), Caroli syndrome (CS), and autosomal recessive polycystic kidney disease (ARPKD). Variants in PKHD1 are responsible for ARPKD and CS with a high inter- and intra-familial phenotypic variability. Rare familial cases of CD had been reported and exceptional cases of CD are associated with PKHD1 variants. In a family of three siblings presenting with a wide spectrum of severity of DPM, we performed whole exome sequencing and identified two PKHD1 compound heterozygous variants (c.10444G>A; p.Arg3482Cys and c.5521C>T; p.Glu1841Lys), segregating with the symptoms. Two compound heterozygous PKHD1 variants, including one hypomorphic variant, were identified in two other familial cases of DPM with at least one patient presenting with CD. This report widens the phenotypic variability of PKHD1 variants to VMC, and others hepatic bile ducts malformations with inconstant renal phenotype in adults and highlights the important intra-familial phenotypic variability. It also showed that PKHD1 might be a major gene for CD. This work adds an example of the contribution of exome sequencing, not only in the discovery of new genes but also in expanding the phenotypic spectrum of well-known disease-associated genes, using reverse phenotyping. © 2015 Wiley Periodicals, Inc.
INTRODUCTION
Ductal plate formation is a key event in porto-biliary embryologic development. Ductal plate malformations (DPM) are characterized by the persistence of embryonic biliary structures and result from remodelling abnormalities of the embryonic ductal plate [Huppert, 2011; Raynaud et al., 2011]. DPM results in congenital diseases that might belong to a phenotypic continuum including from mild to severe presentations, Von Meyenburg complexes (VMC), Caroli disease (CD), Caroli syndrome (CS), and autosomal recessive polycystic disease (ARPKD). VMC is a rare entity, consisting of small, and usually benign multiple hepatic nodular cystic lesions. CD is a rare disease characterized by intrahepatic bile ducts dilatations, first described by Jacques Caroli in 1958 and its association with congenital hepatic fibrosis (CHF) defines CS [Caroli et al., 1958]. Complications of CD are mainly frequent episodes of cholangitis but also liver abscesses, intra- and extra-hepatic lithiasis, and rare cases of cholangiocarcinoma [Büscher et al., 2014]. ARPKD is a congenital disease of the kidney and the liver, belonging to the group of ciliopathies, characterized by fusiform dilatations of the renal collecting ducts, constantly associated with CHF, and often with CS in older patients [Ward et al., 2002; Rossetti et al., 2003; Sgro et al., 2004; Büscher et al., 2014]. Thirty percent of ARPKD patients died during the neonatal and perinatal periods [Bergmann et al., 2005]. The remaining patients develop symptoms during childhood with both renal and hepatic presentations. Only few patients are diagnosed at the adult age mostly with liver-related complications in association with mild kidney disease [Furu et al., 2003; Gunay-Aygun et al., 2010].
PKHD1 is mapped on chromosome 6 and encodes fibrocystin, which is expressed in the primary cilium of renal epithelial cells and bile ducts. Numerous variants associated with ARPKD have been identified in this gene, most of which are unique to single families [Bergmann et al., 2003; Gunay-Aygun et al., 2010].
Rare familial reports with autosomal recessive and autosomal dominant isolated CD have been reported in the literature [Hunter et al., 1966; Turnberg et al., 1968; Höglund et al., 1989; Yoshizawa et al., 1992; Tsuchida et al., 1995; Wu et al., 2002; Benoit et al., 2003; Gunay-Aygun et al., 2013]. In the index cases, CD was usually diagnosed by its complications, mainly cholangitis and hepatic lithiasis, and the other familial cases were diagnosed by systematic hepatic screening. These families were not all screened for variants in PKHD1, but its first implications in isolated CD have been suggested, although not highlighted, in 2003 [Rossetti et al., 2003]. It has now been demonstrated in 2014, by the identification of compound heterozygous PKHD1 variants by exome sequencing in two twins with isolated CD in one case, and CD with renal cysts in the other case [Hao et al., 2014].
Using the same approach, we report here that compound heterozygous PKHD1 variants with at least one hypopmorphic variant can cause several DPM from ARPKD to VMC.
MATERIALS AND METHODS
Patients
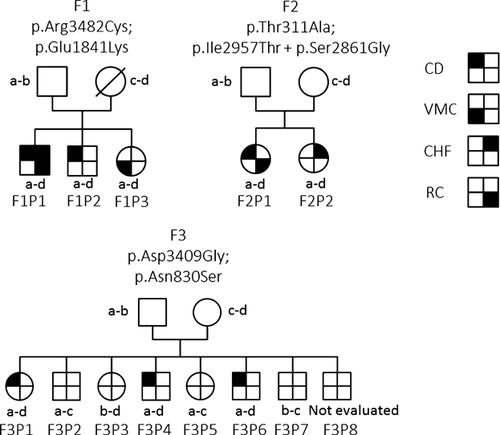
Family 1 was recruited in the clinic with the intent to identify a gene responsible for DPM in a context of recurrence in siblings with three presentations of various degree of the spectrum, including CD and VMC (Table I). Families 2 and 3 were recruited from the literature and a national collaborative call. Family trees are presented on Figure 1. All three families are non-consanguineous. Available phenotypic data were reviewed, and when missing, both renal and hepatic imagery and biology was performed. Estimated glomerular filtration rate (eGFR) were calculated following the Modification of Diet in Renal Disease (MDRD) Study equation.
| F1P1 | F1P2 | F1P3 | F2P1 | F2P2 | F3P1 | F3P4 | F3P6 | |
|---|---|---|---|---|---|---|---|---|
| Age at diagnosis (years) | 7 | 32 | 43 | 29 | NA | 53 | 19 | 20 |
| Diagnosis circumstances | Portal hypertension | Hepatic colic | Systematic screening | Esophageal varices bleeding | Systematic screening | Systematic screening | Cholangitis | Cholangitis |
| Hepatic symptoms | CS | CD | VMC | CD | None | CD | CD | CD |
| Renal symptoms | ARPKD and renal insufficiency | Renal lithiasis | None | Renal cysts with reduced cortico-medullary differenciation | None | None | None | None |
| Variants | c.10444C>T; p.Arg3482Cys | c.10444C>T; p.Arg3482Cys | c.10444C>T; p.Arg3482Cys | c.931A>G; p.Thr311Ala | c.931A>G; p.Thr311Ala | c.10226G>A; p.Asp3409Gly | c.10226G>A; p.Asp3409Gly | c.10226G>A; p.Asp3409Gly |
| c.5521C>T; p.Glu1841Lys | c.5521C>T; p.Glu1841Lys | c.5521C>T; p.Glu1841Lys | c.8870T>C; p.Ile2957Thr + c.8581A>G; p.Ser2861Gly | c.8870T>C; p.Ile2957Thr + c.8581A>G; p.Ser2861Gly | c.2489A>G; p.Asn830Ser | c.2489A>G; p.Asn830Ser | c.2489A>G; p.Asn830Ser |
- Reference sequence (NM_138694.3); CS, Caroli syndrome; CD, Caroli disease; VMC, Von Meyenburg complex; CHF, congenital hepatic fibrosis.
Family 1
F1P1
This patient is the second of three children born from non-consanguineous parents. At 7 years of age, an asymptomatic enlargement of the liver and spleen was discovered fortuitously, and led to investigations. Portal hypertension complicated with esophageal varices and hypersplenism were discovered. Hepatocellular and renal functions were normal. Liver biopsies were performed and revealed dilatations of the intrahepatic bile ducts and fibrosis of the portal spaces. Because hepatic fibrosis could be associated with renal abnormalities, a renal biopsy was performed and revealed dilatations of renal tubules, renal cysts, and tubulo-interstitial fibrosis. The diagnosis of biliary fibroangiomatosis and juvenile renal cystic disease was raised at that time. Regular follow-up during childhood revealed a stabilisation of the disease. At 27 years of age, he presented a first episode of biliary colic. A hepatic magnetic resonance imagery (MRI) was performed and revealed several choledochal stones, a dilatation of intra-hepatic biliary tree and portal vein thrombosis (Fig. 2A). This led to the diagnosis of CS. Renal function was moderately impaired (eGFR: 65 ml/min/1.73 m2). Thereafter, the patient presented seven episodes of biliary stones and cholangitis, and one of pancreatitis, leading to the indication of hepatic transplantation. A renal scan showed renal cysts within the superficial medullar (Fig. 2B). The patient underwent hepatic transplantation at 31 years of age. A second liver transplantation was performed two months later because of liver failure due to hepatic artery thrombosis. During the follow-up, the patient presented with diabetes and a more important renal insufficiency both being side effects of immunosuppressive treatments. He also presented with chronic pancreatitis caused by pancreatic stones. The diagnosis of CS as a manifestation of ARPKD was raised lately, after molecular diagnosis.
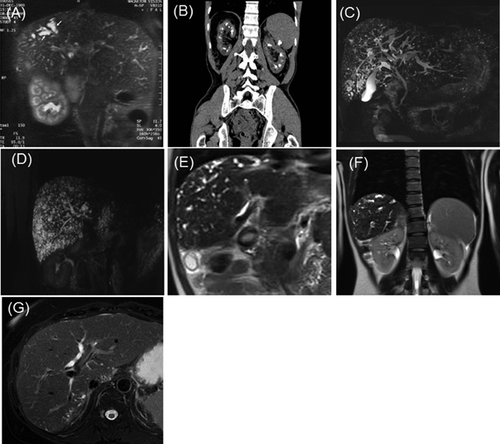
F1P2
This patient presented at 35 years of age with two episodes of renal lithiasis and at 37 years of age, with two episodes of biliary colic with an important cytolysis and cholestasis (ASAT: nine times the normal, ALAT: four times the normal and GammaGT: nine times the normal) (ASAT: 330 UI/L (N:15–37), ALAT: 290 UI/L (N:21–72) and GammaGT: 700 UI/L (N:15–85). Hepatic ultrasound and MRI revealed dilatations of bile ducts without CHF leading to the diagnosis of CD (Fig. 2C). No morphological renal abnormality was detected. Several months later he presented with a pancreatitis and was treated with ursodeoxycholic acid to reduce the formation of biliary lithiasis (GammaGT: 78 UI/L, alcaline phosphatase: 79 UI/L (N:50–136), bilirubinemia: 7 μmol/L (N: 3–17), ASAT: 19 UI/L, ALAT: 45 UI/L, serum creatinine: 61 μmol/L (N:53–115). eGFR was 120 ml/min/1.73 m2 (creatininemia: 92 μmol/L).
F1P3
This patient underwent systematic screening at 43 years of age since her two brothers presented with congenital DPM. A bile ducts MRI revealed hepatic lesions compatible with biliary hamartomas known as VMC and no renal abnormality was detected (Fig. 2D). Hepato-cellular functions were normal except for an isolated elevation of the gammaGT [ASAT: 21 U/L (N < 32), ALAT: 20 U/L (N < 33), GammaGT: 101 U/L (N: 5–36)].
The father of the three siblings had a normal bile ducts MRI, while no information was available for the mother who died from a cancer.
Family 2
F2P1
This female patient presented at 29 years of age with anaemia secondary to esophageal varices bleeding complicating portal hypertension. A hepatic MRI revealed intra-hepatic biliary duct dilatations and a portal vein cavernoma, whose origin remained unknown (Fig. 2E). Liver histology excluded fibrosis. This led to the diagnosis of CD. A renal screening revealed normal biology but reduced cortico-medullary differentiation, thick renal cortical parenchyma and a cyst in both kidneys (Fig. 2F).
F2P2
This female patient is the sister of F2P1. Hepatic and renal US scan were normal as well as biology, with absent intra-hepatic biliary duct dilatations. No liver biopsy was performed to date.
Family 3
Clinical description of the brothers F3P4 and F3P6 had already been published [Benoit et al., 2003]. These two patients are issued from a large family of eight children. CD has been diagnosed in both patients at age 19 and 20, respectively, following recurrent or unique episodes of cholangitis. At the last follow-up, their renal functions were, respectively, at 44 and 41 years of age normal (eGFR: 104 ml/min/1,73 m2) and subnormal (80 ml/min/1,73 m2). Liver functions were also subnormal (Respectively, ASAT: 34 and 32 UI/L (N: 17–59), ALAT: 31 and 59 UI/L (N: 21–72) and GammaGT: 104 and 264 UI/L (N: 12–43). After familial genetic screening, their asymptomatic sister (F3P1) underwent hepatic MRI that suggested a mild presentation of CD with bile duct dilatations limited to the 1st liver segment and to the periphery of the 6th and 7th liver segments and normal kidneys (Fig. 2G and H). Renal function was within normal range (eGFR: 94 ml/min/1,73 m2) as well as liver functions (ASAT: 19 UI/L (N: 14–36), ALAT: 23 UI/L (N: 9–52) and GammaGT: 19 UI/L (N: 15–73). No liver biopsy was performed.
Exome Sequencing
Genomic DNA of subject F1P1 was extracted from whole-blood using standard procedures. Exome capture and sequencing were performed at Integragen (Evry, France) from 3 µg of genomic DNA using the SureSelect Human All Exon V4+UTR kit (Agilent) and a HiSeq 2000 (Illumina) according to the manufacturer's recommendations for paired-end 76 bp reads. Reads were aligned to the human genome reference sequence (GRCh37/hg19) with the Burrows–Wheeler Aligner (BWA, v0.6.2) and potential duplicate paired-end reads were marked using Picard 1.77 [Li et al., 2009]. The Genome Analysis Toolkit (GATK) 2.6-4 was used for base quality score recalibration, indel realignment, and variant discovery [DePristo et al., 2011]. Variants were annotated with SeattleSeq SNP Annotation (http://snp.gs.washington.edu/SeattleSeqAnnotation138/). Rare variants were identified by focusing on protein-altering and splice-site changes present at a frequency below 0.2% in dbSNP 135 and the NHLBI Exome Sequencing Project Exome Variant Server, (http://evs.gs.washington.edu/EVS/), and found in less than 1% of 259 other local exomes. Candidate genes consistent with an autosomal recessive mode of inheritance were systematically identified based on the presence of at least one rare homozygous or two heterozygous variants.
PKHD1 Sequencing
After the identification of PKHD1 variants by exome sequencing in patient F1P1, segregation analysis in all available relatives in family F1 and variant screening in families F2 and F3 were performed by capillary-based sequencing. The 67 PKHD1 exons were amplified. PCR products were purified using 96 wells filtration plates (NucleoFast 96 PCR, Macherey Nagel) and directly sequenced, using Big Dye Terminator (Applied Biosystem, Foster City, CA). The reaction products were purified with BigDye Xterminator Purification Kit (Applied) and loaded on an ABI3730 XL capillary sequencer with POP-7 gel matrix. Data were compared to the reference sequence (RefSeq NM_138694.3) using SeqScape v2.5 software (Applied Biosystem).
The pathogenicity of these variants was evaluated by familial segregation, frequencies reported in Exome Variant Server and in variant specific databases (the Human Genome Mutation Database, http://www.hgmd.org/), a PKHD1 specific database (http://www.humgen.rwth-aachen.de/) and pathogenicity prediction tools for missense variants (Polyphen 2 and Condel) [Adzhubei et al., 2010; González-Pérez and López-Bigas, 2011].
RESULTS
Exome sequencing on a sample of DNA from patient F1P1 generated over 10.5 gigabases of mappable sequence with a median sequencing depth provided for RefSeq coding exons and splice junctions of 77×. The depth of coverage of RefSeq coding bases and splice junctions was of 92% for at least ten reads and 84% for at least twenty reads calculated with the Genome Analysis Toolkit Depth of Coverage tool. After filtering, among the 10 602 non-synonymous, splice site, and indels variants, 284 rare variant remained. Heterozygous missense variants in seven genes were identified (Supplementary Table SI). Given that the phenotype of F1P1 overlapped with that of ARPKD and using reverse phenotyping [Hennekam and Biesecker, 2012], we focused on the two heterozygous variants of the PKHD1 gene: c.10444C>T (p.Arg3482Cys) and c.5521G>A (p.Glu1841Lys). These two variants were also found in his siblings F1P2 and F1P3. Only one variant was found in the father, no DNA from the mother was available.
Sanger sequencing of patients F2P1 and F2P2 revealed two heterozygous variants in the PKHD1 gene: c.931A>G (p.Thr311Ala) inherited from the mother and c.8870T>C (p.Ile2957Thr) inherited from the father, together with a known polymorphism: c.8581A>G (p.Ser2861Gly).
Sanger sequencing in patients F3P1, F3P4, and F3P6 also revealed two heterozygous variants: c.10226A>G (p.Asp3409Gly) inherited from the father and c.2489A>G (p.Asn830Ser) inherited from the mother. Four asymptomatic siblings had only one or none of these variants, the last (F3P8) could not have been evaluated.
Pathogenicity of each variant was argued in Table II.
| Nomenclature (nucleotides) | Nomenclature (aminoacids) | Frequencies in EVS | Present in HGMD | PKHD1 specific database | Polyphen2/HumVar | Condel | ||
|---|---|---|---|---|---|---|---|---|
| F1 | c.10444C>T | p.Arg3482Cys | 1/13500 | Reported | Pathogenic | 0.828 | Possibly damaging | Deleterious |
| F1 | c.5521G>A | p.Glu1841Lys | Not reported | Absent | Unknown | 0.048 | Benign | Neutral |
| F2 | c.931A>G | p.Thr311Ala | Not reported | Absent | Unknown | 0.998 | Probably damaging | Deleterious |
| F2 | c.8870T>C | p.Ile2957Thr | Not reported | Reported | Pathogenic | 0.910 | Probably damaging | Deleterious |
| F2 | c.8581A>G | p.Ser2861Gly | 25/12981 | Reported | Pathogenic, Probably pathogenic | 0.028 | Benign | Neutral |
| F3 | c.10226A>G | p.Asp3409Gly | Not reported | Absent | Unknown | 0.859 | Possibly damaging | Neutral |
| F3 | c.2489A>G | p.Asn830Ser | Not reported | Absent | Polymorphism | 0.005 | Benign | Neutral |
- Reference sequence (RefSeq NM_138694.3); EVS, Exome Variant Server; HGMD, Human Gene Mutation Database.
DISCUSSION
This report, together with the recent literature, demonstrated that PKHD1 is a gene responsible for isolated CD, and also highlights the important range of clinical manifestations of PKHD1 variants. Indeed, the unique phenotype of family one range from VMC to ARPKD, including CD. The use of exome sequencing and reverse phenotyping permitted to explain with success this familial liver phenotype.
Given that the major gene responsible for familial CD cases was not known and the association between PKHD1 and CD had only been rarely reported [Ward et al., 2002; Rossetti et al., 2003; Sgro et al., 2004], we gathered two other familial cases with isolated CD in at least one patient. Later, the association between CD and PKHD1 was specifically highlighted by Hao and collaborators [Hao et al., 2014]. Two variants, one truncating (c.2341C>T, p.Arg781*) and one missense (c.2507T>C, p.Val836Ala) with an unclear impairment on protein function were identified by exome sequencing in two twins with CD.
Interestingly, hepatic conditions associated with ARPKD and PKHD1 variants had been recently studied and reviewed [Gunay-Aygun et al., 2013; Büscher et al., 2014]. Gunay-Aygun et al. showed that 70% of ARPKD patients presented with biliary abnormalities and that there was no correlation between the severity of kidney and hepatic diseases [Gunay-Aygun et al., 2013]. Liver disease could be evidenced on follow-up in all diagnostic groups but was particularly prevalent in those diagnosed later in life. In particular, out of the series of 78 patients, only 10 patients had isolated CHF with normal renal imaging studies (n = 5), one simple renal cyst (n = 1) or no information regarding renal phenotype (n = 4). Rosseti et al. screened for PKHD1 variants using denaturing high performance liquid chromatography 47 pedigrees with ARPKD of various severity and 14 pedigrees with predominant hepatic manifestations including CHF and/or CD, associated or not with kidney manifestations. They identified PKHD1 variants in 5/14 families with predominant hepatic manifestations (32%), but only two had isolated CD and no evidence of renal disease. The first pedigree comprised a unique 36-year-old female with the c.10364delC; p.Ser3455*, and c.10402A>G; p.Ile3468Val variants. The second pedigree comprised male and female siblings with only one identified variant: c.5895_5896insA; p.Leu1966Thrfs*4 [Rossetti et al., 2003]. Unfortunately, clinical information was only mentioned in a table and no details were available. Of interest, several mouse models of ARPKD replicate the findings found in human since they developed severe malformations of intrahepatic bile ducts including bile duct proliferation with progressive cyst formation and associated periportal fibrosis but morphologically and functionally normal kidneys [Moser et al., 2005; Gallagher et al., 2008].
Important intra-familial variability of the clinical expression of ARPKD was shown in 20% of familial cases with heterogeneous hepatic and renal abnormalities and relies on the type of PKHD1 variants [Kaplan et al., 1988; Deget et al., 1995; Bergmann et al., 2005; Adeva et al., 2006; Gunay-Aygun et al., 2013]. Two families are particularly of interest and reflect the importance of modifier genes as well as possible environmental influences.[Adeva et al., 2006] Indeed, one patient was diagnosed at 1 year of age with ARPKD whereas his sister, carrying the same PKHD1 variants, did not display any symptoms at 51 years. In another patient, the diagnosis of ARPKD was raised at 9 months on the association of hepatomegaly and enlarged kidneys, and his two sibs carrying the same PKHD1 alleles, only had a few renal cysts diagnosed by systematic screening. Our observations confirm the intra-familial phenotypic variability associated with variants of PKHD1 on a cohort of patients with familial DPM, but also highlight the importance of familial screening of asymptomatic siblings after the identification of an index case.
ARPKD, CD, and CS are caused by a wide range of PKHD1 variants [Ward et al., 2002; Hao et al., 2014]. No genotype-phenotype correlations were found regarding the location of variant and the severity of hepatic and renal symptoms in classical ARPKD [Gunay-Aygun et al., 2013, 2010]. However, a genotype-phenotype correlation was found regarding the types of variants [Bergmann et al., 2003]. Indeed, patients with two truncating variants displayed a very severe phenotype with death in the perinatal period. Survival with two variants in the PKHD1 gene require that at least one of the two variants is a missense variant [Bergmann et al., 2003]. Whereas the role of truncating variants of PKHD1 is clear, interpreting the pathogenicity of missense variant within PKHD1 remains an issue, especially the role of the association between hypomorphic variants and clearly pathogenic variants [Bergmann et al., 2005; Gunay-Aygun et al., 2010]. In our report, all families have one pathogenic or possibly pathogenic missense variant, associated to another missense variant considered as hypomorphic in two families (p.Glu1841Lys in F1 and p.Asn830Ser in F3). Of notice, Gunnay-Aygun et al., showed that heterozygous carrier of PKHD1 variants could present with mild asymptomatic hepatic or renal malformations in some cases [Gunay-Aygun et al., 2011]. This hypothesis of compound heterozygosity appear as the best hypothesis as compared with PKHD1 heterozygosity only since the index cases were all symptomatic, the variants fully segregated with the symptoms in the reported families, and similar results were found in the family recently reported by Hao [Hao et al., 2014]. The association of missense/hypomorphic variants should be the clue to explain this milder end of the PKHD1 phenotypic spectrum. In such mild phenotypes, systematic hepatic and renal screening, with at least biology testing and imagery, appears mandatory in asymptomatic siblings of affected patients, in order to determine at risk patients that should beneficiate from regular follow-up.
In conclusion, the present report (i) demonstrates that PKHD1 can be considered as a major gene for CD; (ii) gives another example of reverse phenotyping following exome sequencing investigations; (iii) widens the spectrum of PKHD1 variants from the benign VMC to the severe form of ARPKD and highlights that isolated liver phenotypes could be assigned to PKHD1 variants and not only in association with renal manifestations, with a probable underdiagnosis; and (iv) further highlights the phenotypic variability and suspects the role of hypomorphic variants.