Microduplication 3q13.2q13.31 identified in a male with dysmorphic features and multiple congenital anomalies
Abstract
Constitutional microdeletions affecting 3q13.2q13.31 are rare and attempts for genotype–phenotype correlations have only recently been made in a cohort of 28 patients. The major phenotypic features of this rare syndrome are hypotonia, developmental delay, and facial anomalies. In this study, we report on a male infant with a novel reciprocal 3.671 Mb microduplication at the genomic region 3q13.2q13.31 associated with dysmorphic features and multiple congenital anomalies. The current patient was investigated by high-resolution array comparative genomic hybridization (aCGH). This is the first report of a microduplication 3q13.2q13.31 that shares a lot of common clinical features with those carrying the microdeletion. The 3q13.2q13.31 duplicated region in our patient contains nine dosage sensitive genes, amongst them the genes ATG3, CCDC80, KIAA2018, NAA50, ZDHHC23, DRD3, ZBTB20, GAP43, LSAMP. As it is the case for many other well-described reciprocal deletion/duplication syndromes, some have very different clinical features (Williams–Beuren deletion syndrome, WBS/WBS triplication) [Somerville et al. (2005); N Engl J Med 353:1694–1701], while others share similar phenotypic features (22q11.2 microdeletion/microduplication) [Portnoi (2009); Eur J Med Genet 52:88–93]. In conclusion, we describe the main phenotypic features of a possibly novel microduplication 3q13.2q13.31 syndrome. Additionally five of the dosage-sensitive genes and BOC gene are suggested to be responsible for the main phenotypic features. Evaluation of multiple patients with the microduplication is needed for full delineation of this syndrome. © 2013 Wiley Periodicals, Inc.
INTRODUCTION
Deletions at the proximal long arm of chromosome 3 (3q11q23) are extremely rare and due to variability in the deletion breakpoints, they present with variable phenotypes. A recent report of 14 new well-characterized patients with deletions of 3q12.3q21.3 [Molin et al., 2012] and a review of another 14 previously published cases identify the common phenotypic features and also define the shortest region of overlap (SRO), 0.6 Mb in size, located at 3q13.31. The SRO is shared by 24 out of the 28 microdeletion patients and contains five genes (DRD3, ZNF80, TIGIT, MIR568, ZBTB20) two of which (DRD3 and ZBTB20) are strong candidate genes for developmental delay (DD) [Lawson-Yuen et al., 2006; Shimojima et al., 2009; Molin et al., 2012].
We describe the first case of a reciprocal microduplication to the newly described 3q13.31 microdeletion syndrome in a newborn male with dysmorphic features and multiple major congenital anomalies, sharing common features with the newly recognized 3q13.31 microdeletion syndrome and presenting with a 3.671 Mb duplication of 3q13.2q13.31, identified by array comparative genomic hybridization (aCGH). The DD candidate genes (DRD3 and ZBTB20) and other dosage sensitive genes (ATG3, CCDC80, KIAA2018, NAA50, ZDHHC23, GAP43, LSAMP) are included in the duplicated interval [Huang et al., 2010].
PATIENTS AND METHODS
Clinical Report
This patient was the first child of a nonconsanguineous couple. The mother was obese; otherwise both parents had a normal clinical phenotype and cognitive function. During the pregnancy, the mother was on modified diet due to an abnormal glucose tolerance test. She underwent an amniocentesis for chromosome analysis due to advanced maternal age showing a normal male karyotype 46, XY. Second trimester level II ultrasound scan (US) and color Doppler showed intrauterine growth restriction (IUGR), oligohydramnios as well as increased uterine artery resistance.
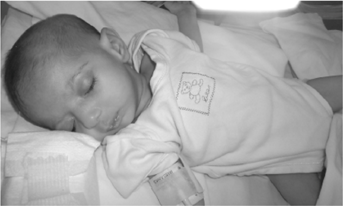
The infant was delivered by elective cesarean due to breech presentation at 38 + 3 weeks. At birth he was slightly hypotonic but did not require resuscitation. Birth weight was 2,460g (3rd centile), length 45.5 cm (3rd centile), and head circumference 32.2 cm (3rd centile). Clinical examination revealed hypotonia, a small for gestational age (SGA) male newborn with moderate hypertelorism, microretrognathia, a wide cleft palate, overlapping toes, a deep sacral dimple, and glandular hypospadias (Fig. 1, Table I).
| Patient phenotypes | Duplication patient (this report) | 24 Cases with microdeletion overlapping with SROa |
|---|---|---|
| Age | 1y | Birth—19y6m |
| Sex | Male | 15/25 |
| Birth weight (g) | 2,460 (3rd centile) | Normal |
| Birth length (cm) | 45.5 (3rd centile) | Normal |
| Birth OFC (cm) | 32.2 (3rd centile) | 4/11 have >85th |
| Weight (g) | 5,600 (≪3rd centile) | 50th → 99th |
| Length (cm) | 67 (≪3rd centile) | 10/21 have >85th |
| OFC (cm) | 41.5 (≪3rd centile) | 9/20 have >85th |
| CNS malformations/brain | Small left subependymal cyst cluster formation | 3 ventriculomegaly, 5 agenesis of corpus callossum, 1 alobar holoprosencephaly |
| DD | Mild | 19/21 |
| Speech delay | ? (Too young) | 15/17 |
| Behavior problems | ? (Too young) | 8 autism/hyperactivity, 1 epilepsy |
| Hypotonia | + | 12/15 |
| Skull abnormalities | 2 dolichocephaly, 2 plagiocephaly, 3 brachycephaly | |
| Prominent/broad forehead | Broad forehead | 10/13 |
| Strabism | − | 6/14 |
| Myopia | − | 4/8 |
| Ptosis | − | 4/11 |
| Epicanthal folds | − | 8/14 |
| Hypertelorism | Moderate | 7/17 |
| Downslanting palpebral fissures | − | 7/13 |
| Short philtrum | − | 6/6 |
| High palate | − | 7/10 |
| Ears | − | 5 large/15, 4 low set/15 |
| Abnormal external genitalia (for males) | Glandular hypospadias | 11/15 |
| Skeletal abnormalities | − | 16/24 |
| Other malformations | Wide cleft palate, 7-mm perimembranous ventricular septal defect, pulmonary hypertension, patent foramen ovale, PDA, mild tricuspidal insufficiency, right pulmonary artery stenosis, severe gastroesophageal reflux, mild hearing impairment | Midline cleft lip and palate, PDA, hearing impairment, nystagmus, tetralogy of Fallot, crowded teeth, kidney abnormalities, uretal anomaly, malformed sacrum, spina bifida, ileal atresia, etc. |
| Other dysmorphic features | Microretrognathia, overlapping toes, deep sacral dimple, sits independently but does not stand or walk | Pointed chin, flat or prominent nasal bridge, broad nasal bridge, anteverted nares, everted or thin lips, short palpebral fissures, short sternum, long philtrum, skin anomalies, sits independently but does not stand or walk, dysarthric speech |
- PDA, patent ductus arteriosus.
- a Molin et al. [2012].
Apneic episodes in the first postnatal days of life were fully investigated. Brain ultrasound showed first a left subependymal hemorrhage and thereafter a small left subependymal cyst cluster formation.
Echocardiography performed due to a grade II systolic murmur, heard at the 3rd postnatal day, identified a 7-mm perimembranous ventricular septal defect and pulmonary hypertension, patent foramen ovale, patent ductus arteriosus, mild tricuspidal insufficiency, and right pulmonary artery stenosis. He was treated initially with digoxin and diuretics and thereafter ductus arteriosus ligation and pulmonary artery banding procedure.
Although fed with the help of a Haberman nipple for cleft palate, he presented with feeding problems, severe gastroesophageal reflux and growth failure. Fluoroscopy of the upper gastrointestinal tract and abdominal ultrasound showed no abnormalities. Nasogastric feedings were used until the age of 6 months.
Furthermore, ophthalmological examination was normal and audiological assessment showed abnormal transient otoacoustic emissions (TOAEs) on the left. Additional evaluation with auditory brainstem responses (ABR) showed normal hearing at high frequencies with 20 dBHL threshold on the left ear and mixed hearing loss at high frequencies with 60 dBHL threshold on the right ear.
At the age of 3 months he showed head lag and remarkable truncal hypotonia. Brain MRI showed no remarkable findings and cysts seen during the immediate postnatal period were not present anymore.
At the age of 1 years old, his body weight was 5,600g, height 67 cm, head circumference 41.5 cm (all ≪3rd centile). He had mild neurodevelopmental delay. He could sit without support but not pull to standing position; he could not creep and crawl or walk unassisted. He could grasp objects. He could not pronounce repetitive consonant sounds but he was responding to the sound of his name. His feeding difficulties improved.
The presence of multiple minor and major congenital anomalies led to the further genetic investigation. With the informed consent of the parents, aCGH was performed. The research was approved by the University of Athens Ethics committee.
Methods
Genomic DNA was isolated from blood leukocytes. Array CGH was performed using high-resolution Agilent 4x180K arrays, following the manufacturer's protocol and our previous publication [Tzetis et al., 2012]. The parents refused testing but on clinical examination showed normal cognitive function and did not present with any phenotypic abnormalities.
RESULTS
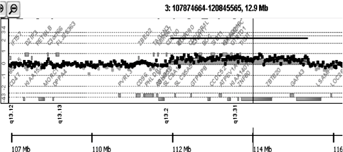
High-resolution aCGH revealed only one pathogenic finding (duplication 3q13.2q13.31; 3.67 Mb; 112152600–115823438; hg19) (Fig. 2), with the rest of the findings being benign (Supplementary Table I—see Supporting Information Online).
DISCUSSION
Interstitial deletions of proximal long arm of chromosome 3 (3q12q21) have been associated with a wide range of phenotypes. Deletions of 3q12q21 are rare, with 28 cases published to this date plus a few cases reported in databases such as ISCA and DECIPHER (https://www.iscaconsortium.org/, http://www.sanger.ac.uk/PostGenomics/Decipher) [Shimojima et al., 2009; Molin et al., 2012]. The patient described here shares the following phenotypic features with patients bearing the 3q13.31 microdeletion: hypertelorism, cleft palate, abnormal genitalia (hypospadias), DD, congenital heart defect and hypotonia (Fig. 1, Table I), as well as overlapping toes and mild peripheral sensorineural hearing impairment [Lawson-Yuen et al., 2006; Shimojima et al., 2009; Molin et al., 2012]. However, our patient also presented with phenotypic features that were not present in patients with the 3q13.31 microdeletion such as microretrognathia, intrauterine and postnatal growth retardation and feeding difficulties while the majority of patients reported with the reciprocal microdeletion syndrome showed postnatal overgrowth [Molin et al., 2012]. Reports of feeding difficulties, dyspnea and tachypnea were reported for three patients with the 13q13.2 deletion [Shimojima et al., 2009]. Since our patient is only 12 months old, we cannot determine whether he may develop autistic features associated with the reciprocal microdeletion [Molin et al., 2012] (Table I).
The genomic duplicated region 3q13.2q13.31 present in our patient contains 26 RefSeq genes, five of which (DRD3, ZNF80, TIGIT, MIR568, ZBTB20) are contained within the SRO shared with the 24 of 28 microdeletion patients [Molin et al., 2012; Shimojima et al., 2009]. Additionally the region contains haploinsufficient (dosage sensitive) genes (ATG3, CCDC80, KIAA2018, NAA50, ZDHHC23, DRD3, ZBTB20, GAP43, LSAMP) with haploinsufficiency (HI) scores <50% [Huang et al., 2010]. The presence of dosage sensitive genes in the region supports the theory of overexpression phenotypes that resemble the underexpression ones [Conrad and Antonarakis, 2007]. This hypothesis could explain the common features of our patient with those previously described with deletions in the same genomic region. The same observations have been made for microduplication 22q11.2, complementary to the 22q11.2 deletion syndrome and also 16p13.3 microduplication which is complementary to 16p13.3 microdeletion causing Rubinstein–Taybi syndrome [Portnoi, 2009; Thienpont et al., 2010]. These findings indicate that normal gene dosage might be restricted not only by a lower threshold but also by an upper threshold, giving an explanation of the same organs and functions often involved in these reciprocal deletion and duplication syndromes [Thienpont et al., 2010]. However as more reciprocal deletion/duplication syndromes are characterized, several examples of contrasting phenotypic effects have also been reported (proximal 16q11.2 microdeletion and microduplication, 7q11.23 in Williams–Beuren (WBS) deletion and triplication syndromes, NSD1 deletions causing Sotos syndrome and reciprocal 5q35 duplication) [Somerville et al., 2005; Rosenfeld et al., 2013].
The 3q13.2q13.31 genomic region contains genes highly expressed in the brain that might be subject to dosage imbalance. Two genes are particularly interesting with respect to DD; DRD3 the dopamine receptor D3 gene expressed in the limbic area of the brain and ZBTB20 expressed in the developing hippocampus neurons [Hoenicka et al., 2007; Beninger and Banasikowski, 2008; Molin et al., 2012]. Other candidate DD genes contained in the region are LSAMP and GAP43, both dosage sensitive. LSAMP encodes the limbic system associated membrane protein and studies in mice and humans have supported its involvement in behavioral and neuropsychiatric disorders [Catania et al., 2008]. GAP43 (neuron growth-associated protein 43) is involved in neurite outgrowth, neurotransmission and synaptic plasticity. Recently it has also been implicated as a candidate gene for autism [Zaccaria et al., 2010].
The common characteristic of all patients with the microdeletion was muscular hypotonia, a feature also observed in our patient. Two candidate genes could be responsible for this clinical feature. One of them is BOC which is involved in the interaction and differentiation of the myogenic cells [Kang et al., 2002, 2003]. Another candidate gene could be CCDC80 (coiled-Coil Domain containing 80), which is involved in axon pathfinding, guiding motor neuron axons towards skeletal muscles forming thereafter synaptic contacts [Brusegan et al., 2012]. Loss of the homologous gene in zebrafish embryos induces motility issues without impairment of myogenesis. Expression of CCDC80 in mice white adipose tissue additionally suggests a role of the gene in the regulation of body weight and metabolism [Brusegan et al., 2012]. One could hypothesize that overexpression of CCDC80 could be responsible for the low growth parameters observed in our case with the 3q13.2q13.31 duplication, in contrast to the overgrowth shown in the majority of patients with the reciprocal microdeletion [Shimojima et al., 2009; Molin et al., 2012]. Another dosage sensitive gene, ZBTB20, has also been implicated in growth and metabolism, therefore causing the postnatal overgrowth phenotype seen in patients with the reciprocal microdeletion and perhaps its overexpression could be responsible for the undergrowth phenotype in our case [Sutherland et al., 2009].
CONCLUSION
In the era of aCGH our patient might represent a new microduplication syndrome which is complementary to the novel microdeletion syndrome of the same genomic region recently described [Shimojima et al., 2009; Molin et al., 2012]. To further support this hypothesis more cases with this novel microduplication need to be studied.




