Co-occurrence of Prader–Willi and Sotos syndromes†
How to Cite this Article: Okamoto N, Akimaru N, Matsuda K, Suzuki Y, Shimojima K, Yamamoto T. 2010. Co-occurrence of Prader–Willi and Sotos syndromes. Am J Med Genet Part A 152A:2110–2111.
Abstract
A patient with atypical phenotypes of Prader–Willi syndrome (PWS) was subjected to investigate genomic copy numbers by microarray-based comparative genomic hybridization analysis. Severe developmental delay, relative macrocephaly, protruding forehead, cardiac anomalies, and hydronephrosis were atypical for PWS. Concurrent deletions of 15q11-13 and 5q35 regions were revealed and identified as paternally derived. The sizes and locations of the two deletions were typical for both deletions. Although each deletion independently contributed to the clinical features, developmental disturbance was very severe, suggesting combined effects. This is the first report of co-occurrence of PWS and STS. The co-occurrence of two syndromes is likely incidental. © 2010 Wiley-Liss, Inc.
INTRODUCTION
Prader–Willi syndrome (PWS; OMIM #176270) is caused by deficiency of paternally expressed imprinted transcripts within chromosome 15q11-q13 [Ledbetter et al., 1981]. It is characterized by obesity, hypotonia, hypogonadism, and behavioral abnormalities [Holm et al., 1993]. Most paternal PWS deletions are bracketed by recurrent breakpoints (BP)1 or BP2 and BP3. Preturbed expression of genes including SNURF–SNRPN and multiple small nucleolar RNAs (snoRNAs) are associated with the clinical manifestations of PWS, but the specific contributions of individual genes are under investigation. Recent analysis revealed that deficiency of HBII-85 snoRNAs causes the key characteristics of the PWS phenotype, although some atypical features suggest that other genes in the region may make more subtle phenotypic contributions [Sahoo et al., 2008].
Sotos syndrome (STS; OMIM#117550) is an overgrowth syndrome characterized by pre- and postnatal overgrowth, macrocephaly, developmental delay, advanced bone age, and a distinctive face including frontal bossing, frontal sparseness of hair, hypertelorism, downslanting palpebral fissures, and pointed chin. Haploinsufficiency of the NSD1 gene due to 5q35 microdeletions or intragenic mutations causes STS [Kurotaki et al., 2002]. Miyake et al. 2003 observed that microdeletions in STS are mostly of paternal origin. Common deletion breakpoints were located at two flanking low copy repeats (LCR), implying that non-allelic homologous recombination (NAHR) between LCRs is the major mechanism for the common deletion in STS [Kurotaki et al., 2005; Visser et al., 2005]. Central nervous system anomalies, cardiovascular and urogenital symptoms are more frequent in the microdeletion group [Nagai et al., 2003].
In this study, a patient with atypical phenotypes of PWS was subjected to investigate genomic copy numbers by microarray-based comparative genomic hybridization (aCGH) analysis. Concurrent deletions of 15q11-13 and 5q35 regions were detected and identified as paternally derived. Although each deletion independently contributed to the clinical features, growth and developmental disturbance were very severe, suggesting combined effects. This is the first report of co-occurrence of PWS and STS.
CLINICAL REPORT
A 14-year-old male propositus is the first-born child of healthy and non-consanguineous parents. After uncomplicated pregnancy, he was born at 39 weeks of gestation by induced delivery with overgrowth of length with 53 cm (90th centile).
His birth weight was within a normal limit as 3,010 g (25th centile). He was the first child of a 26-year-old mother and a 30-year-old father. Since cardiac murmur was found at birth, he was transferred to the neonatal intensive care unit and ventricular septal defect (VSD), atrial septal defect (ASD), and patent ductus arteriosus (PDA) were revealed by echocardiography. Micropenis and bilateral cryptorchidism were noticed. He had severe hypotonia and feeding difficulties in the early infantile period. Until his sucking improved at 6 months old, nasal tube feeding was required. Ultrasonography revealed bilateral vesicoureteral reflux and hydronephrosis. He showed a severe developmental delay with head control at 1 year of age and sitting alone at 6 years of age. He had generalized seizures at age 6 years. Electroencephalography revealed sporadic spikes at that time. Brain MRI showed no significant findings. He developed progressive obesity, as his weight was 10.0 kg (75th centile) at 9 months old of age and 12.4 kg (95th centile) at 1 year old of age. Conventional G-band chromosome analysis showed a normal male karyotype, and subsequent conventional FISH analysis for SNRPN revealed a deletion, indicating a diagnosis of PWS. In spite of that, relative macrocephaly, protruding forehead, frontal baldness, and mild overgrowth were atypical for phenotypic features of PWS (Fig. 1A). Although he was interested in food, hyperphagia was not prominent because of his restricted locomotive abilities. Gradually, his height SD scores decreased (Fig. 2). Partial growth hormone deficiency was found by endocrinological studies. When he was 14 years of age his bone age was measured at the 11-year-old level. His parents did not choose GH replacement therapy.
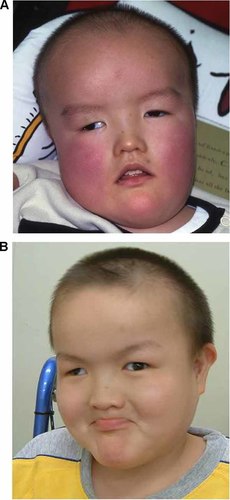
Facial appearance of the patient at 6 years old (A) and 14 years old (B).
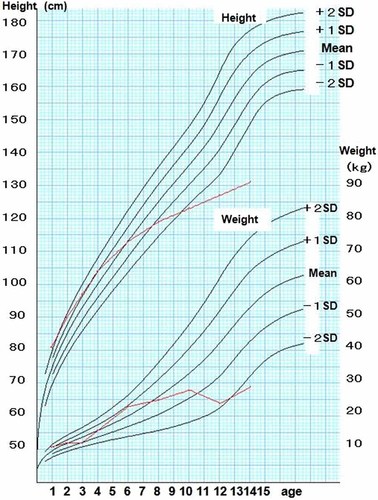
Growth curve of the patient. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
When we examined the patient at the age of 14 years, he showed severe mental retardation without vocalized words, muscular hypotonia, hypopigmentation, scoliosis, and distinctive facial features including protruding forehead; strabismus; hypertelorism; downslanting palpebral fissures; epicanthal folds; full cheeks; microstomia with downturned corners of the mouth; small hands with tapering fingers; and small feet (Fig. 1B). A wheel chair was required for him because his hip joint was unstable and he could not stand alone. His intelligent quotient (IQ) was measured by Kyoto Scale of Psychological Development as below 10. He was a calm and friendly boy. His interest in food became obvious, but self-injurious behaviors such as skin picking were not observed. Behavioral problems associated with STS including autistic spectrum disorder, hyperactivity, and aggression were not present. His weight was 29 kg (<3rd centile), and his length was 132 cm (<3rd centile) (Fig. 2). His head circumference was mean for his age. A comparison of typical features of PWS and STS and their clinical presentation in the patient are shown (Table I).
| Prader–Willi | Sotos | Current patient | |
|---|---|---|---|
| Hypotonia | + | + | ++ |
| Mental delay | + | + | ++ |
| Hypopigmentation | + | − | + |
| Prominent forehead | − | + | + |
| Strabismus | + | + | ++ |
| Over growth | − | + | − |
| Growth delay | + | − | ++ |
| Obesity | + | − | + |
| Epilepsy | − | + | + |
| Congenital heart disease | − | + | + |
| Scoliosis | + | + | ++ |
| Hydronephrosis | − | + | + |
| Hypogonadism | + | − | + |
- +, common features; ++, prominent manifestations.
MATERIALS AND METHODS
After obtaining informed consents based on a permission approved by the institution's ethical committee, peripheral blood samples were obtained from the patient and his parents. Genomic DNAs were extracted using the QIAquick DNA extraction kit (QIAgen, Valencia, CA).
Based on the hypothesis that the patient might have an atypically larger deletion of chromosome 15 or have additional chromosomal aberrations, aCGH analysis was performed using the Human Genome CGH Microarray 60K (Agilent Technologies, Santa Clara, CA) as described previously [Shimojima et al., 2009].
Metaphase nuclei were prepared from peripheral blood lymphocytes by mean of standard methods and used for FISH analysis with human BAC clones selected from the UCSC genome browser (http://www.genome.ucsc.edu) as described elsewhere [Shimojima et al., 2009]. Physical positions refer to the March 2006 human reference sequence (NCBI Build 36.1).
Microsatellite marker analysis was performed using the ABI Prism Linkage Mapping Set with D15S1002 and analyzed by GeneMapper (Applied Biosystems, Foster City, CA). In the deletion region of STS, no marker was available for the ABI Prism Linkage Mapping Set. Thus, the single-nucleotide polymorphisms (SNP) typing was carried out. From the STS deletion region of 5q35, eight SNPs, IMS-JST038690, IMS-JST087588, IMS-JST087589, IMS-JST183486, IMS-JST172005, IMS-JST073857, IMS-JST087921, and IMS-JST087922, were selected using in silico library, Japanese Single Nucleotide Polymorphisms (JSNP) database (http://snp.ims.u-tokyo.ac.jp/index.html). Allelic types were analyzed by PCR-direct sequencing method using the BigDye terminator (Applied Biosystems, Foster City, CA).
RESULTS
By aCGH analysis, loss of the genomic copy numbers was identified in the region of 15q11.2, which is responsible and typical for PWS (Fig. 3A). The concurrent deletion was identified in the region of 5q35, which is also responsible and typical for STS (Fig. 3B). FISH analyses confirmed the deletion of both regions (Fig. 4). There were no deletions of PWS region and STS region in both parents indicating de novo occurrence (data not shown).
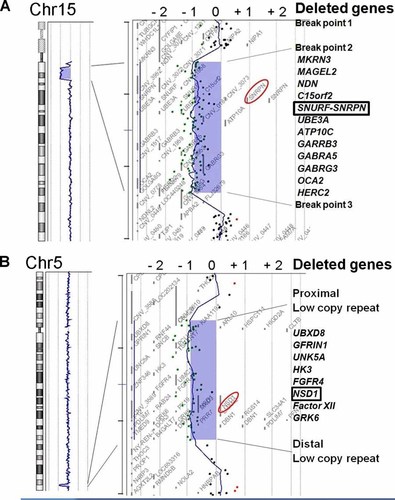
aCGH profiles of the patient shown by CGH Analytics in Chromosome view (left) and Gene view (right). A: Typical deletion of PWS region including SNRPN is shown. B: Typical STS deletion including NSD1 is indicated. The horizontal axis indicates the log 2 ratio of the genomic copy number. The blue rectangles indicate the regions containing copy number aberrations. The aberration areas are expanded in Gene view (right). The dots indicate the locations and the corresponding log 2 ratios of the probes. The red circles emphasize SNRPN and NSD1.
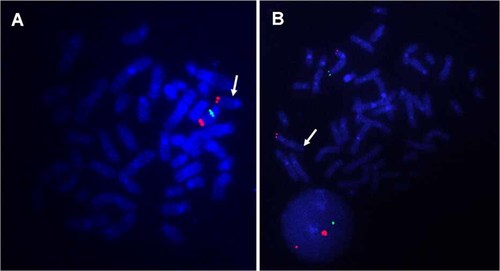
FISH analysis to confirm the chromosomal deletion. A: One of the green signals covering SNRPN, RP11-1071C22 (15q11.2; 22601976–22822028), was deleted. Two red signals are the markers of chr15, RP11-48A4 (15q26.3; 99433829–99587322). B: One of the green signals covering NSD1, RP11-99N22 (5q35.2–5q35.3; 176474586–176655375), was deleted, whereas two red labeled RP11-94J21 (5p15.33; 1377471–1540913) signals were confirmed in all cells. Physical positions are referred to NCBI Build 36.1. White arrows indicate abnormal chromosomes in each FISH image.
To confirm the parental origin of both deletions, polymorphic markers were analyzed in the patient and his parents. Regarding the 15q11.2 region, the patient showed an only allele with 112-bp common to his mother, indicating the deletion of paternal allele (Fig. 5A). Among eight analyzed SNPs, only IMS-JST183486 was informative. The patient showed hemizyogous of T at the SNP position, whereas the father and the mother showed homozygous of A and T, respectively (Fig. 5B). From the result, we concluded that both deletions were derived from the paternal allele.
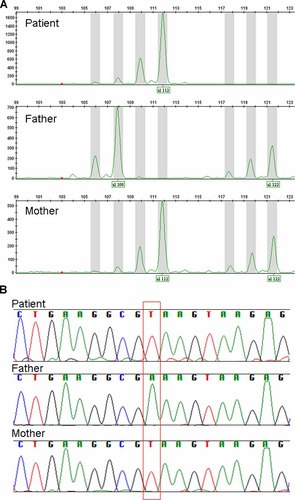
Molecular analysis of the patient's family. A: GeneMapper analysis using D15S1002. The patient shows only one allele with 112-bp common with his mother, indicating the paternal deletion. B: SNPs analysis of IMS-JST183486. The patient's SNP type as T is only common with his mother, indicating the deletion of paternal allele.
DISCUSSION
Initially, the patient was diagnosed as PWS due to severe hypotonia, hypopigmentation, hypoplastic genitalia, and small hands and feet. It was supported by hyperphagia and obesity which later developed. However, his facial features including relative macrocephaly, protruding forehead, frontal baldness, strabismus, downslanting palpebral fissures, and pointed chin were atypical for PWS. He also showed congenital cardiac anomalies, hydronephrosis, and epilepsy, which are rare findings in PWS. Severe hypotonia and severe developmental delay were also atypical for PWS. This was the reason why we analyzed genomic copy numbers.
To the best of our knowledge, this is the first report of co-occurrence of PWS and STS. Translocation between chromosomes 5 and 15 was excluded by G-banded analysis. Array CGH demonstrated that the sizes and locations of the two deletions were typical for both syndromes. Both of the deletions were derived from the paternal chromosome. We suspect that co-occurrence of two deletions is incidental.
His growth curve showed an interesting pattern. He showed overgrowth in the infantile period. Gradually, his growth velocity decreased. Now he shows severe growth deficiency. Although we understand that haploinsufficiency of NSD1 might lead to height gain and patients with STS show advanced bone age, his growth deficiency was worse compared with standard PWS patients and his bone age was delayed [Nagai et al., 2000]. Growth hormone deficiency and severe scoliosis may explain his growth deficiency. We posit that each deletion contributed independently to the features. Severe growth and developmental delay might be explained by the combined effects of PWS and STS.
There are some reports of concurrent chromosomal aberrations in the same patients [Shimojima et al., 2009]. The result of this study indicates that there may be more frequent co-occurrences of two more deletions than what we think. When a patient shows atypical or overlapping features regardless of a previously established diagnosis, we would recommend investigation of whole genomic copy numbers by aCGH.
Acknowledgements
We thank the family for their co-operation. This study was supported by the Health and Labour Research Grants in 2009 by Ministry of Health, Labour and Welfare in Japan.




