Distal 3p deletion syndrome: Detailed molecular cytogenetic and clinical characterization of three small distal deletions and review†
How to cite this article: Malmgren H, Sahlén S, Wide K, Lundvall M, Blennow E. 2007. Distal 3p deletion syndrome: Detailed molecular cytogenetic and clinical characterization of three small distal deletions and review. Am J Med Genet Part A 143A:2143–2149.
Abstract
The distal 3p deletion syndrome is characterized by developmental delay, low birth weight and growth retardation, micro- and brachycephaly, ptosis, long philtrum, micrognathia, and low set ears. We have used FISH and BACs in order to map three 3p deletions in detail at the molecular level. The deletions were 10.2–11 Mb in size and encompassed 47–51 known genes, including the VHL gene. One of the deletions was interstitial, with an intact 3p telomere. In nine previously published patients with 3p deletions, the size of the deletion was estimated using molecular or molecular cytogenetic techniques. The genotype, including genes of interest, and the phenotype of these cases are compared and discussed. The localization of the proximal breakpoint in one of our patients suggests that the previously identified critical region for heart defects may be narrowed down, now containing three candidate genes. We can also conclude that deletion of the gene ATP2B2 alone is not enough to cause hearing impairment, which is frequently found in patients with 3p deletion. This is the third reported case with an interstitial deletion of distal 3p. © 2007 Wiley-Liss, Inc.
INTRODUCTION
Since the first case was described by Verjaal and De Nef 1978, less than 30 patients with distal 3p deletions have been reported. Despite the low number of patients, a recognizable phenotype including developmental delay, low birth weight and growth retardation, micro- and brachycephaly, ptosis, long philtrum, micrognathia, and low set ears, has become evident. In nine cases, the extent of the deletion has been more thoroughly investigated using molecular cytogenetic techniques or RFLP/microsatellite analyses [Mowrey et al., 1993; Phipps et al., 1994; Drumheller et al., 1996; Green et al., 2000; Cargile et al., 2002]. Cell lines are available from some of the patients, and these have been included in several of the studies (Table I). A majority of the patients have been shown to have terminal deletions with breakpoints in 3p25.3. However, at the molecular level the breakpoints vary, and for some less consistent features like congenital heart defects (CHD), candidate genes have been identified. The variation in the breakpoints also includes the von Hippel–Lindau tumor suppressor gene (VHL), located in 3p25.3. So far, no patients with deletion of the VHL gene have been reported to have hemangioblastomas or other signs of von Hippel–Lindau disease, but most patients are still very young and VHL usually presents later in life. Distal, interstitial 3p deletions have only been reported in two cases, the smallest one comprised approximately 4.5 Mb and identified the hitherto smallest region of overlap for the distal 3p deletion syndrome [Cargile et al., 2002]. In addition, a patient with many of the physical features of distal 3p deletion syndrome and a de novo reciprocal translocation between chromosomes 3 and 10 was recently reported [Fernandez et al., 2004]. The breakpoint on chromosome 3 disrupted the Contactin 4 (CNTN4) gene, which is situated in 3p26.3. CNTN4 was therefore suggested to have an important role in the development of distal 3p deletion syndrome and the CNS.
| Patient reference(s) | CUMG3.1 P2 | DZ GM10922 P3 | CUMG3.4 P5 | Patient 1 | Patient 2 | Patient 3 LD GM10985 P8 | CUMG3.10 P10 | DR | TL | — | TR | — |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Paper(s) where the patient is included |
Ramer et al. 1989, Mowrey et al. 1993, Phipps et al. 1994, Drumheller et al. 1996, Green et al. 2000 |
Present study | Present study |
Present study Ramer et al. 1989, Mowrey et al. 1993, Phipps et al. 1994, Drumheller et al. 1996, Green et al. 2000 |
Drumheller et al. 1996 |
Angeloni et al. 1999 |
Mowrey et al. 1993 |
Cargile et al. 2002 |
||||
| Sex | M | M | F | F | F | F | F | M | F | M | M | M |
| Birth weight (g) | 2,350 | 2,570 | 2,300 | 2,390 | 2,270 | 2,150 | 2,060 | 2,320 | 2,523 | 3,450 | NR | 2,695 |
| Developmental delay 12/12 | + | + | + | + | + | + | + | + | + | + | + | + |
| Growth retardation 12/12 | + | + | + | + | + | + | + | +/− | + | + | + | + |
| Feeding problems 6/11 | NR | + | + | − | + | + | + | NR | NR | NR | + | NR |
| Hypotonia 10/11 | + | + | + | − | + | + | + | NR | + | + | + | + |
| Microcephaly 8/11 | + | + | + | − | − | + | + | − | + | − | + | + |
| Brachycephaly 6/8 | − | − | + | ? | + | + | + | NR | NR | NR | + | + |
| Micrognathia 11/12 | + | + | + | + | + | + | + | − | + | + | + | + |
| Blepharophimosis 6/7 | NR | + | NR | − | + | + | + | NR | NR | NR | + | + |
| Hypertelorism/telecanthus 10/12 | + | + | − | + | + | + | − | + | + | + | + | + |
| Hooded eyelids/epicanthus 8/8 | NR | + | NR | + | + | + | + | NR | + | NR | + | + |
| Ptosis 8/11 | + | + | + | − | − | + | + | − | + | NR | + | + |
| Periorbital fullness 6/8 | − | + | NR | + | + | + | − | NR | NR | NR | + | + |
| Flat nasal bridge 7/8 | + | − | NR | + | + | + | + | NR | NR | NR | + | + |
| Anteverted nares 9/10 | + | + | NR | + | + | + | + | + | − | NR | + | + |
| Low set/round ears 10/11 | + | + | NR | +? | + | + | + | + | + | + | + | + |
| Preauricular pits/tags 2/11 | + | − | + | − | − | − | − | + | − | NR | − | − |
| Long philtrum 10/11 | + | + | NR | + | + | + | + | + | + | − | + | + |
| Hearing impairment 4/7 | NR | NR | NR | − | + | + | − | − | + | + | NR | NR |
| Heart malformation 3/12 | + | + | + | − | − | − | − | − | − | − | − | − |
| Renal malformation 3/6 | NR | NR | − | − | + | + | NR | NR | NR | − | + | NR |
| Postaxial polydactyly 3/12 | − | − | − | + Toe | − | + | − | + | − | − | − | − |
| Sacral skin dimple 4/5 | NR | NR | NR | − | + | NR | + | + | NR | NR | NR | + |
- NR, not reported.
- The patients are listed according to the estimated location of the proximal breakpoint, starting with the most proximal breakpoint to the left. Patients from the present study in bold.
We have investigated three patients with distal 3p deletion syndrome and, to the best of our knowledge, the third reported interstitial deletion of distal 3p. The extent of the deletions was outlined in detail using FISH-mapping and BACs, and has been compared to the previously reported cases.
MATERIALS AND METHODS
The study was approved by the Research Ethics Committee at Karolinska Institutet.
Patient 1
This girl was born as the second child to healthy, non-consanguineous parents. The pregnancy was uncomplicated, with delivery at gestational age 36 weeks and 5 days. The birth weight was 2,390 g, birth length 44 cm, and head circumference 31.5 cm. She was observed at a neonatal ward during the first week due to tachypnea and minor sucking difficulties. At 2 months of age, infantile spasm-like seizures were noted, but EEG at that time was normal. Neurological examination revealed a slight head lag, but otherwise normal, symmetric muscular tonus. The psychomotor development was delayed and dysmorphic features were noted at 4 months of age: up-slanting eyes, epicanthus, a flat nasal bridge, a broad, up-turned nose, and a long philtrum. In addition, she had a short neck, a large anterior fontanel and an extra digit on the left foot. At 16 months of age, primary hypothyroidism, and malrotation of the colon were found. She had also developed moderate scoliosis. No heart or kidney malformations could be detected. She learned to sit at 1 year of age, but never acquired intentional grasping. She had a delayed and poor eye contact and no speech. A CT scan showed a small brain with normal morphology.
At the age of 5 years, Patient 1 had developed stereotypic hand movements, air bloating, teeth grinding and screaming attacks, and she had a high threshold to pain. She had muscular hypotonia except in the feet, which were small with dystonic posturing. She was considered to fulfill the criteria for congenital Rett syndrome, but no MECP2 mutation could be found, and she was reported as a patient with a congenital variant Rett syndrome in 1999 [Wahlström et al., 1999]. SPECT showed decreased perfusion patterns bitemporally and a minor decrease in perfusion in the frontal lobes and around the third ventricle. Laboratory investigations for metabolic disorders were negative.
At 15 years of age her developmental status is essentially unchanged. Menarche was at 14 years of age. She is small, −3 SD for length and weight, and −2 SD for head circumference. She has normal vision, but avoids eye contact. The hearing is probably normal, as she reacts to low sounds. The tendon reflexes are normal, but there is slightly increased and varying tonus in the lower extremities. Patient 1 has epilepsy with several attacks per month, mainly generalized attacks, and has over time developed focal epileptiform activity on the EEG, with background activity changing from normal to generalized severe slowing. A recent examination of the eyes and ultrasonography of the abdomen showed no signs of hemangioblastoma.
Patient 2
This girl was born as the second child to a 31-year-old mother and a 34-year-old father. The pregnancy was complicated by polyhydramniosis and an amniocentesis was performed in pregnancy week 33. Chromosome analysis revealed a distal deletion of the short arm of chromosome 3. The patient was born in pregnancy week 35, with a birth weight of 2,270 g, birth length 44 cm, and head circumference 31.5 cm. Apart from physiological jaundice treated with phototherapy, the neonatal period was normal. Mild dysmorphic features could be noted: a flat nasal bridge, a broad, up-turned nose, low set ears, a long philtrum and thin lips, epicanthus, periorbital fullness, and a small chin (Fig. 1). In addition, there was a sacral dimple. No heart or kidney malformations, normal hands and feet and a normal muscular tone were found. An MRI scan of the brain was performed at 3 months of age and showed probable delayed myelinization. At the age of 5 months, Patient 2 had mild muscular hypotonia and a head lag, she could lift her head from the floor, but made no attempts to roll over. She gave normal eye contact and made different sounds. The tendon reflexes were normal. At 9 months of age she could roll over, but was still slightly hypotonic.
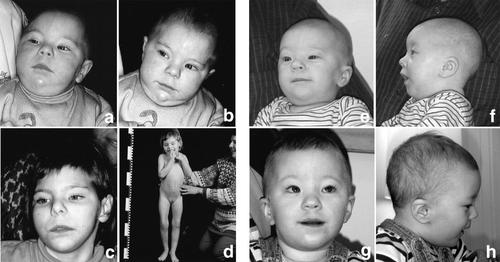
Pictures of Patient 1 at 10 months (A,B) and 6 years of age (C,D), and of Patient 2 at 5 months (E,F), and 18 months of age (G,H). Note the similar facial features, for example, epicanthus, periorbital fullness, a short, up-turned nose, the shape of the mouth, a long philtrum, and a small chin.
At her present age of 2 years and 8 months, Patient 2 is still hypotonic, and has no voluntary ability to move by herself. She has developed epilepsy with general tonic clonic seizures, especially at night. In addition, there are severe sleep disturbances. Another main problem has been feeding difficulties and vomiting, and she has been treated for gastro-esophageal reflux. The feeding difficulties have diminished, but she could not chew at the age of 18 months. She has a hearing impairment. Ultrasonography has revealed a horseshoe malformation of one kidney. At birth the weight, length, and head circumference were all at −2 SD, corrected for gestational age, and she has continued to follow −2 SD for most parameters.
Patient 3
This female patient was first reported by Ramer et al. 1989, but has been included in four additional studies [Mowrey et al., 1993; Phipps et al., 1994; Drumheller et al., 1996; Green et al., 2000]. We refer to the first paper for a description of the phenotype, which was followed up to 8 months of age. A lymphoblastoid cell line (GM10985) from this patient was obtained from the NIGMS (Human Genetic Mutant Cell Repository Coriell Institute for Medical Research, Camden, NJ).
Cytogenetic Studies
Metaphase slides were prepared from amniotic fluid, peripheral blood cultures, and lymphoblastoid cell lines according to standard procedures. Chromosome analysis was performed according to routine methods using both QFQ- and GTG-banding techniques, including high resolution banding.
FISH
BACs/PACs mapped to human chromosomes 3 were chosen based on their mapped position. The BAC clones that define the breakpoints are shown in Figure 3. They were ordered from CHORI or Sanger and delivered as bacterial LB agar stab cultures and the clones were isolated and prepared according to standard protocols. The BAC/PAC probes, labeled with FITC or SpectrumOrange by nick translation, were hybridized on metaphases from the patients together with a centromeric probe from chromosome 3 (CEP3, Vysis, Downers Groove, IL) according to standard protocols. The signals were visualized using a Zeiss Axiophot fluorescence microscope equipped with a cooled CCD-camera (Cool SNAP HQ, Photometrics Ltd, Tuscon, AZ), controlled by a Macintosh Quadra 950 computer. Gray scale images were captured, pseudo colored and merged using the SmartCapture software (Digital Scientific, Cambridge, UK). At least 10 metaphases per hybridization were analyzed. The databases http://www.ensembl.org and http://www.ncbi.nlm.nih.gov were used for picking out relevant BAC-clones, identification of genes, and physical mapping.
RESULTS
Cytogenetic Analysis
In Patient 1, a distal deletion of 3p was demonstrated, and the karyotype thus reported as 46,XX,del(3)(p25.2). Chromosome analysis of amniotic fluid as well as peripheral blood from Patient 2 showed a deletion of the short arm of chromosome 3, which was suspected to be interstitial. The karyotype was reported as 46,XX,del(3)(p25.3p26.3) (Fig. 2). The parental karyotypes were normal in both cases. The karyotype in Patient 3 was described as 46,XX,del(3)(p25) [Ramer et al., 1989]. No parental karyotype was reported.
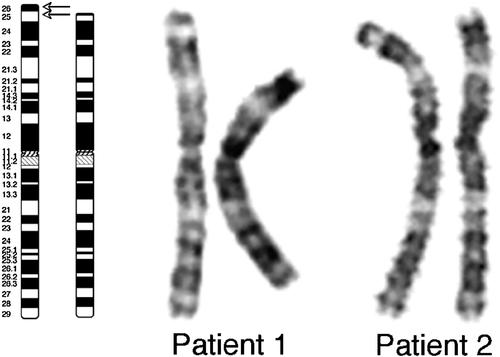
Ideograms of the normal and deleted chromosome 3 are shown to the left. The lower arrow indicates the proximal breakpoint in all three patients. The upper arrow indicates the distal breakpoint in Patient 2. G-banded chromosomes of the normal (right) and deleted (left) copies of chromosome 3 in Patients 1 and 2 are shown to the right.
FISH Mapping
The breakpoint in Patient 1
A 3p subtelomere probe was previously used and reported as deleted [Wahlström et al., 1999]. Further mapping showed that the BAC clone RP11-627C1 was deleted, while RP11-109J15 was retained. According to present information in Ensembl, BAC clone RP11-627C1 is located at 10.79–10.96 Mb, and RP11-109J15 at 10.99–11.14 Mb, thus only separated by 30 kb. A previously identified candidate gene for heart defects was therefore deleted in this patient (SLC6A11).
The distal breakpoint in Patient 2
The BAC clone RP11-114K9 (0.33–0.52 Mb) was deleted, while the partially overlapping telomeric BAC clone RP11-306H5 (containing marker D3S4559, D3S2359, and D3S3938) as well as TelVysion 3p (Vysis) was retained, but with a diminished fluorescent signal. Therefore, part of this telomeric clone is probably deleted in the patient. It covers the CHL1 (CALL) gene which is most likely disrupted by the breakpoint. The CHL1 gene is the most telomeric gene known on 3p and has been suggested a candidate gene for mental retardation in 3p deletion patients [Frints et al., 2003].
The proximal breakpoint in Patient 2
The BAC clone RP11-327H17 (10.61–10.71 Mb) was deleted and RP11-627C1 (10.79–10.96 Mb) was retained. The clone between these two, RP11-193K15 (10.69–10.87 Mb), was retained but gave a weaker signal on the deleted chromosome, thus the breakpoint occurred within this clone. No genes appear to be located in the breakpoint region. The proximal breakpoint in Patient 2 is 0.2 Mb more distal, but very close to the breakpoint in Patient 1. Only one gene differs, which is retained in Patient 2 (SLC6A11). RP11-327H17 contains the markers D3S3381, D3S3107, D3S3611, D3S2416, D3S3594, D3S1675, D3S3601, D3S587, and RP11-627C1 contains the markers D3S3589 and D3S3327.
The interstitial deletion in Patient 2 is approximately 10 Mb in size, and encompasses all genes from CHL1, which is disrupted, down to the ATP2B2 gene. In total, 47 known genes are deleted.
The breakpoint in Patient 3
Detailed mapping of the breakpoint in the patient previously described as LD [Ramer et al., 1989; Mowrey et al., 1993; Phipps et al., 1994; Drumheller et al., 1996; Green et al., 2000], was performed using the cell line GM10985 (Human Genetic Mutant Cell Repository Coriell Institute for Medical Research, Camden, NJ). The BAC clone RP11-1082A18 (10.35–10.52 Mb) is retained in Patient 3. The clone RP11-438J1 (10.14–10.35 Mb) showed a weak signal on one of the chromosome 3, implicating that the breakpoint is located within this clone. This confirms previous results that showed that marker D3S1317 is deleted in this patient [Drumheller et al., 1996]. The breakpoint in this patient was located 0.6 Mb distal to the breakpoint in Patient 2, which means that the genes ATPB2, GHRL, SEC13L1, TATDN2 are retained in Patient 3, while deleted in Patient 2. The breakpoint in Patient 3 is most likely disrupting the gene IRAK2.
DISCUSSION
The distal 3p deletion syndrome is a contiguous gene syndrome that is rather well defined, despite the low number of reported cases. We have performed an extensive investigation of three distal 3p deletions, of which one was interstitial. The size of the deletions was estimated to 10.2–11 Mb and encompassed 47–51 known genes. A comparison with previously characterized distal 3p deletion cases is shown in Table I and Figure 3 [Tolmie et al., 1986; Schwyzer et al., 1987; Ramer et al., 1989; Tazelaar et al., 1991; Mowrey et al., 1993; Phipps et al., 1994; Drumheller et al., 1996; Angeloni et al., 1999; Green et al., 2000; Cargile et al., 2002]. Although similar at the cytogenetic level, no common breakpoint has been found, and the deletions vary in size from patient to patient. There are some segmental duplications on distal 3p, but none of them directly at the breakpoints of our patients. Therefore, we have no indications that the deletions are caused by non-allelic homologous recombination between low-copy repeats, similar to 22q11 and 7q11.23 deletions. Most of the characteristic symptoms, like growth retardation, feeding problems, hypotonia, microcephaly and the very characteristic facial features (Fig. 1), are present in practically all patients, including the one with the smallest interstitial deletion [Cargile et al., 2002]. The gene(s) responsible for the main clinical picture are therefore most likely located within this minimal region of overlap (MRO). The MRO contains 12 genes (CNTN4, IL5RA, TRNT1, CRBN, LRRN1, SUMF1, SETMAR, ITPR1, BHLHB2, EDEM1, ARLB, and Q26EZ6), some of which are expressed in brain and have been suggested as candidates for the mental retardation, for example, LRRN1 and ITPR1. Interestingly, a case with a reciprocal translocation disrupting the CNTN4 gene, close to the distal boundary of the MRO, was recently published [Fernandez et al., 2004]. The patient had physical features of the distal 3p deletion syndrome and a causal relationship between CNTN4 disruption and the phenotype was suggested.
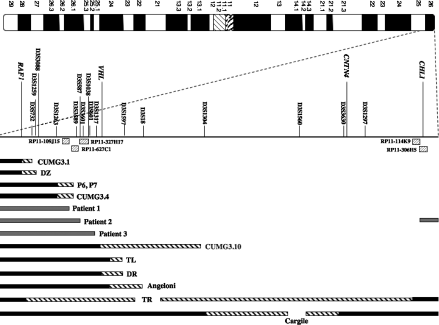
A schematic picture comparing the reported size of the 3p deletions found in the patients included in Table I. Patients P6 and P7 were mapped by Green et al. 2000 but no detailed phenotype was reported, and these patients were therefore not included in Table I. The filled bars show the verified retained regions (gray for the patients included in this study, black for the previously reported cases). The striped parts of the bars indicate the possible breakpoint location, reflecting the varying mapping-resolution of the previously reported deletions. The BAC clones that defined the deletion breakpoints in Patients 1, 2, and 3 are shown as striped boxes. In Patient 1 RP11-109J15 (10.99–11.14 Mb) is retained while RP11-627C1 (10.79–10.96 Mb) is deleted, defining the breakpoint at approximately 10.9 Mb. Patient 2 has an interstitial deletion with the distal breakpoint within the clone RP11-306H5 (0.22–0.34 Mb), disrupting the most distal gene CHL1. The proximal breakpoint is defined by the clone RP11-327H17 (10.61–10.71 Mb) that is deleted, while the adjacent proximal clone RP11-627C1 (10.79–10.96 Mb) is retained. In Patient 3, the breakpoint has occurred within the clone RP11-438J1 (10.14–10.35 Mb). Detailed, updated information regarding clone localization and genes in this region can be found at http://www.ensembl.org.
The deletions in our two new patients resemble the one previously described for Patient 3 (cell line GM10985). The deletion breakpoint in Patient 3 is 0.6 Mb distal to the breakpoint in Patient 2 and disrupts the gene IRAK2, thus four additional genes are deleted in Patient 2, including ATP2B2, GHRL, SEC13L1, and TATDN2. ATP2B2 is the most proximal of these genes and has been suggested as a candidate for hearing impairment [Street et al., 1998]. A mouse animal model has shown that mice with homozygous ATP2B2 mutations are deaf and that heterozygous mice have significant hearing loss. However, this gene is not deleted in Patient 3 and therefore most likely not responsible for the hearing impairment in that particular patient. In addition, Patient 1, CUMG3.1, DZ, and CUMG3.4 have deletions that include ATP2B2, but no reported hearing impairment, and TL as well as the patient reported by Angeloni et al. 1999 have documented hearing problems, but no deletion of the same gene. Indeed, the fact that patients with smaller deletions have hearing problems while those with larger deletions do not, indicates that solely haploinsufficency of ATP2B2 is not enough to cause hearing problems. One may speculate that unmasking of a recessive gene by the deletion, or the influence of more than one gene may be alternative mechanisms giving rise to varying hearing loss in distal 3p deletion patients.
Some, but not all patients with distal 3p deletion syndrome have CHD, and a critical region between D3S3589 and D3S1263 has been suggested [Green et al., 2000]. This interval is approximately 0.65 Mb and contains four candidate genes, SLC6A11, SLC6A1, HRH1, and ATG7. Previously suggested candidates like ATP2B2, Sec13R [Drumheller et al., 1996], and CRELD1 (previously cirrin) [Green et al., 2000] can now be excluded as they have been mapped outside the critical region. The deletions in Patients 2 and 3 do not include the CHD region, while the deletion in Patient 1 involves the most distal gene SLC6A11, and accordingly, as no abnormality of the heart has been found in Patient 1, this finding might further narrow down the candidates and the localization of the critical region for CHD to 0.45 Mb, now containing three genes. SLC6A1 is a neurotransmitter transporter (GABA), HRH1 is a histamine receptor and ATG7 is a gene important for autophagy. Loss of ATG7 leads to neurodegeneration [Komatsu et al., 2006]. Hence, none of these three genes is an obvious candidate for CHD.
Patient 2 has been followed up to 3 years and Patient 1 up to 15 years. As for many chromosomal syndromes, very few adults with distal 3p deletion have been described. The long-term knowledge regarding prognostic aspects of the syndrome is therefore poor. The oldest patient, a 25-year-old woman (the mother of DR, Table I), was of short stature (148 cm), with a slow speech development, academic skills at the 6-year-old level and social skills at the 7- to 8-year-old level. She did not have seizures, but approximately half of the patients older than 2 years have been reported to have problems with epilepsy. Reports of older patients are especially important with regard to deletions of the VHL gene. The long-term consequences in these cases are still unknown and it can only be speculated upon the possible risk for the development of von Hippel–Lindau disease-associated tumors. Until further cases have been documented, patients with VHL deletions must be recommended to undergo the same control program as a standard VHL-patient.
In contrast to all other reports, a woman with a 3p25.3 deletion and a seemingly normal phenotype was reported by Knight et al. 1995. The deletion was transferred to her son who was also reported to be unaffected. The deletion was confirmed using a subtelomere probe, but the detailed extension of the deletion is unfortunately unknown.
In conclusion, we have performed fine mapping of three patients with distal 3p deletions, one of them the hitherto third reported interstitial deletion of distal 3p. The deletions were 10.2–11 Mb in size and encompassed 47–51 known genes, including VHL. The localization of the proximal breakpoint in one patient suggests that the previously identified critical region for heart defects may be narrowed down, and we can also conclude that deletion of the gene ATP2B2 alone is not enough to cause hearing impairment.




