Characterization of terminal chromosome anomalies using multisubtelomere FISH
Abstract
Telomeric repeat sequences (TTAGGG) are known to cap the termini of every human chromosome. Proximal to these repeat sequences are chromosome-specific repeat sequences, which in turn are distal to gene-rich regions. Submicroscopic, subtle, or cryptic abnormalities in these regions can now be investigated using commercial probe sets for all of the chromosome-specific subtelomeric regions of the human genome. Using this technology, previously unidentified genomic imbalance has been found in a proportion of patients with idiopathic developmental delay and learning difficulties. We have used these probe sets to investigate cases with apparently terminal anomalies detected on G-banded chromosome analysis. As a result of such investigations, we have found that 3 (19%) of 16 apparently terminal deletion cases were the result of more complex rearrangements involving other chromosome subtelomeres. The remaining 13 cases contained no chromosome-specific subtelomere repeats on the deleted arm, but in all 16 cases, the TTAGGG telomere repeat cap was present. A further case was investigated where extra material was found in the terminal region of the chromosome 12 short arm, found to represent a complex inversion/duplication/deletion rearrangement. Investigation of all cases with terminal anomalies, including apparently terminal deletions, is likely to uncover further cases involving complex rearrangements and should lead to a greater understanding of the mechanisms by which these abnormalities arise. © 2003 Wiley-Liss, Inc.
INTRODUCTION
Telomeres have a number of essential roles within the cell [McClintock, 1941]. They prevent end-to-end chromosome fusion which would otherwise result in the production of dicentric and ring chromosomes and subsequent instability and chromosome breakage in successive cell cycles. They also provide a ‘buffer zone’ at the chromosome ends, which shortens during every cell cycle due to the nature of semi-conservative replication, preventing the degradation of the middle-repetitive telomere-associated regions and gene-rich regions proximal to the telomeres. In humans, the enzyme telomerase is responsible for telomere maintenance [Morin, 1989], but is thought to be active only in the germ cells and not in somatic cells beyond 16–20 weeks of embryonic development [Wright et al., 1996], although one study [Fan et al., 2000] has described low levels of telomerase in normal human lymphocytes. Therefore, another telomerase-independent mechanism for telomere maintenance may exist in somatic cells. Such a mechanism, involving recombination with a telomere from another chromosome and use of this as a template for telomere extension, using DNA polymerase and the recombination proteins Rad51p and Rad52p, has been postulated [Dunham et al., 2000].
The subtelomeric regions are gene-rich and due to the repeat motifs found in these areas may be particularly prone to rearrangements, which could give rise to viable abnormal phenotype. The commercial subtelomeric probes now available [National Institutes of Health and Institute of Molecular Medicine Collaboration, 1996; Knight and Flint, 2000] are a powerful tool for investigating anomalies involving the distal regions of chromosome arms, and have been used to test for cytogenetically cryptic or submicroscopic abnormalities in patients with learning difficulties and dysmorphism [Warburton et al., 2000; Joyce et al., 2001; Anderlid et al., 2002].
These probes have also been used to investigate a small number of cases of apparently terminal deletions [Horsley et al., 1998; Ballif et al., 2000], and some of these cases were shown to be subtle semi-cryptic rearrangements involving the terminal regions of other chromosomes. The involvement of other chromosomes means that the chromosomal imbalances in such cases may be different from those that were originally supposed and therefore will have a bearing on the phenotype of the individual.
This study describes the results from the investigation of 16 cases of apparently terminal deletions and a further interesting case where G-banded chromosome analysis showed additional material in the distal region of the chromosome 12 short arm.
MATERIALS AND METHODS
Metaphase chromosome preparations were obtained from peripheral blood lymphocytes using standard cytogenetic techniques.
In situ hybridization was carried out using standard procedures. Probes used were as follows. Chromoprobe-T probes and the Multiprobe-T system from Cytocell, Banbury, UK; chromosome-specific subtelomere probes from Q-BIOgene, Livingston, UK; ToTelVysion, probe pair D5S23/EGR1, WHS, STS, KAL, and TEL-ETV6 from Vysis, Inc., Downers Grove; the XIST probe, DXZ1, and the AHT (all human telomere) from Appligene Oncor, Harefield, UK, and human whole chromosome paints from Cambio, Cambridge, UK.
All of the apparently deleted cases were initially tested with the chromosome-specific subtelomere probe pair for the short and long arm of that chromosome. They were then investigated by multisubtelomere FISH using either commercial kits, used according to the manufacturer's instructions, or telomere DNA probes used in a multiwell system previously described [Mackie Ogilvie et al., 1997, 1998]. Other locus-specific probes and whole chromosome paints were used for further investigations. FISH signals were visualized using a Powergene image analysis system (Applied Imaging, Newcastle, UK).
CLINICAL CASES
Sixteen individuals were investigated using multisubtelomere FISH after G-banded chromosome analysis had revealed apparently terminal deletions. These individuals ranged in age from 2 days to 45 years. Eleven had developmental delay or learning difficulties. Ten had other dysmorphic features or congenital abnormalities. One had secondary amenorrhoea but no other apparent abnormality. Two were apparently normal and their chromosomal abnormalities were discovered following investigation of their daughters, one of whom had been referred for possible Turner syndrome or Noonan syndrome and was found to have a maternally inherited deletion of the long arm of chromosome 10, and the other who died in the neonatal period and was found to have a maternally inherited deletion of the short arm of the X chromosome. Two individuals, SW and LS, were referred with a specific request to exclude subtelomere deletions of chromosomes 1q and 2q, respectively, based solely on phenotype. SC had been previously determined to have a mosaic karyotype with one normal female cell line and one female cell line with a terminal deletion of chromosome 2q and further characterization was requested. The phenotypes and initial karyotypes of these cases, reported prior to fluorescence in situ hybridization studies, are summarized in Table I. Blood samples from parents of these individuals were requested in all cases; 12 out of the 16 abnormalities were shown to have arisen de novo, and in the remaining 4 cases, parental blood samples were not available.
| Case | Age at referral | Phenotype | Result of initial G-banded chromosome analysis | |||||
|---|---|---|---|---|---|---|---|---|
| Dev delay learning diff | Craniofacial abnormalities | Hypotonia | Short stature | CHD | Other | |||
| SW | 2 years | + | + | − | − | − | Agenesis of corpus callosum, see text | 46,XX,del(1)(q44) |
| LS | 16 years | + | + | − | − | − | Obesity | 46,XX,del(2)(q37.2) |
| SC | 17 years | + | − | − | − | − | IDDM, autoimmune hypothyroidism | 46,XX,del(2)(q36.3)[19]/46,XX[11] |
| ND | 15 years | + | − | − | − | − | Nasal speech | 46,XY,?del(5)(p15.33) |
| JR | 22 years | + | + | − | + | Apnoea | 46,XX,del(7)(q34) | |
| OS | 2 months | + | + | − | − | − | Widely spaced nipples, hypospadias | 46,XY,del(9)(p23) |
| LA | 7 months | + | + | + | − | − | 46,XX,del(10)(q26.1) | |
| JG | 45 years | − | − | − | − | − | Daughter is 46,XX,del(10)(q?26.1) | 46,XX,del(10)(q26.2) |
| CF-N | 13 days | N/A | − | − | − | − | Folded right ear, imperforate anus | 46,XX,?del(10)(q26.1) |
| CD | 5 years | + | − | − | + | + | Clinodactyly | 46,XY,?del(11)(q24.2) |
| ME | 2 days | N/A | + | − | − | + | Absent kidney, microphthalmia, absent corpus callosum | 46,XX,del(11)(q24.1) |
| DW | 1 year | + | + | − | − | − | Low-set ears | 46,XY,?del(13)(q32) or (q33) |
| RF | 6 years | + | − | − | − | − | Articulation problems | 46,XX,del(18)(p?11.2) |
| RR | 6 months | +(Mild) | + | + | − | − | Proximally placed thumbs, clinodactyly, umbilical hernia | 46,XX,?del(18)(q21.3) |
| AA | 28 years | − | − | − | − | − | Daughter (deceased) was 46,X,del(X)(p22.1) | 46,X,del(X)(p22.13) |
| HW | 18 years | − | − | − | − | − | Secondary amenorrhoea | 46,X,del(X)(q?21.1) |
| CS | 40 years | + | + | − | − | − | Sleep apnea, overweight, etc. (see text) | 46,XX,add(12)(p13.3) |
- Clinical details and results of initial G-banded chromosome analyses.
A further case, CS, was investigated by FISH following discovery of additional material on the short arm of chromosome 12. She was referred at 40 years of age for karyotype analysis and FISH analysis for deletion of chromosome 22q11.2; she had always been considered to have Turner syndrome although she had never been karyotyped. She had had delayed milestones with menstruation starting at 13 years of age but only occurring approximately once per year until she was 26 years old when her periods became monthly. She suffered with sleep apnoea and was overweight with a long columella, high narrow palate, poor dentition, narrow palpable fissures, puffy feet with short toes and nails, and long tapering fingers. G-banded chromosome analysis showed an abnormal female karyotype 46,XX,add(12)(p13.3) with additional material in the short arm of one chromosome 12. Parental karyotypes are unknown.
RESULTS
The 16 cases with apparently terminal deletions were initially investigated by FISH using subtelomere probe pairs specific for the short and the long arms of the apparently deleted chromosomes. In all 16 cases, the probe for the apparently deleted subtelomeric region did not hybridize to the abnormal chromosome. All cases were then investigated by multisubtelomere FISH. In all cases, the signal for the deleted subtelomere probe was again shown to be absent from the abnormal homolog and in 13 of the cases no signal from any other chromosome-specific subtelomere probe was shown to be present at the terminus of the deleted chromosome arm (see Table II). However in the case of three individuals, a hybridization signal from a chromosome-specific subtelomere probe from another chromosome was present at the terminal region of the deleted chromosome arm (see Table II). Multisubtelomere FISH on SW showed a signal corresponding to the chromosome 6 long arm specific subtelomere probe at the terminus of the deleted chromosome 1 long arm. Similarly the same test on LS showed a signal corresponding to the chromosome 5 long arm-specific subtelomere probe at the terminus of the deleted chromosome 2 long arm, and on ME revealed hybridization of the chromosome 4 short arm specific probe at the terminus of the deleted chromosome 11 long arm. All FISH results are summarized in Table II.
| Case | Abnormal chromosome | Multisubtelomere FISH | Results of additional FISH tests on abnormal chromosome | |||
|---|---|---|---|---|---|---|
| Probe set | Short arm probe and result | Long arm probe and result | Additional hybridization on abnormal chromosome | |||
| SW | ?del(1)(q44) | Multiprobe-T | CEB108+ | D1S3739− | D6S2522+ (6q) | |
| LS | ?del(2)(q37.2) | Multiprobe-T | D2S2983+ | D2S2986− | D5S2097+ (5q) | |
| SC | ?del(2)(q36.3) | To TelVysion | yRM1051+ | yRM2112− | None | |
| ND | ?del(5)(p15.3) | To TelVysion | C84C11− | GS35o8+ | None | D5S23+, EGR1+ |
| JR | ?del(7)(q34) | Multiprobe-T | 109A6+ | 2000a5− | None | |
| OS | ?del(9)(p23) | Multiprobe-T | 43N6− | D9S2168+ | None | |
| LA | ?del(10)(q26.1) | Multiprobe-T | D10S2488+ | D10S490− | None | |
| JG | ?del(10)(q26.2) | Multiprobe-T | D10S2488+ | D10S490− | None | |
| CF-N | ?del(10)(q26.1) | To TelVysion | 10PTEL006+ | D10S2490− | None | |
| CD | ?del(11)(q24.2) | Multiprobe-T | D11S2071+ | D11S4974− | None | |
| ME | ?del(11)(q?24.1) | To TelVysion | D11S2071+ | RM2072− | GS10K2+ (4p) | WHS− |
| DW | ?del(13)(q32)or(q33) | Multiprobe-T | N/A | D13S1825− | None | |
| RF | ?del(18)(p?11.21) | To TelVysion | D18S552− | D18S1390+ | None | |
| RR | ?del(18)(q21.3) | Q-Biogene | 52M11+ | D18S3090− | None | |
| AA | ?del(X)(p22.1) | Q-Biogene | DXYS129− | Xqsubtel+ | None | |
| HW | ?del(X)(q?21.2) | Multiprobe-T | CY29+ | C8.1/2− | None | XIST+ |
| CS | add(12)(p13.3) | To TelVysion | 8M16− | RM2196+ | None | WCP12+,TEL-ETV6++ |
In four cases, further FISH investigations were carried out in an attempt to verify breakpoints (see Table II). ND was further investigated by FISH using the probe D5S23 specific for the Cri du chat region in band 5p15.2. This was shown to be present (as was the control probe EGR1 at 5q31) on both homologs of chromosome 5. HW was further investigated by FISH using a probe for XIST at Xq13 on the long arm of the X chromosome and was present on both homologs of the X chromosome. ME was tested with the probe WHS for the Wolf–Hirschhorn locus at 4p16.3, which was shown to be absent from the derivative chromosome 11, although it was present on both homologs of chromosome 4. AA was investigated using probe STS specific for the steroid sulphatase locus and KAL specific for the Kallmann locus both within Xp22.3 and DXZ1 specific for the centromere of the X chromosome. The X centromere probe hybridized to both the deleted X and the normal X chromosomes, but both the STS and the KAL probes hybridized only to the normal X chromosome.
Case CS, which had additional material on the short arm of one homolog of chromosome 12, was initially investigated using whole chromosome paints specific for each of the chromosomes. Only the chromosome 12-specific paint hybridized to the abnormal chromosome 12 and appeared to do so along its entire length. Multisubtelomere FISH showed that none of the chromosome-specific subtelomere probes hybridized to the short arm of the abnormal chromosome 12, although hybridization to all other chromosome arms was normal. However, the use of probe TEL-ETV6, which maps to 12p13, showed that the abnormal chromosome 12 had two discrete TEL-ETV6 signals on the short arm while the normal homolog of chromosome 12 had only the expected one signal. These results and retrospective examination of the G-banding allowed the interpretation of this chromosome abnormality as an inverted duplication with associated distal deletion, as has been described previously on other chromosomes [Jenderny et al., 1998; Bonaglia et al., 2000]. The final karyotype was: 46,XX,inv dup del(12)(qter → p13.3::p13.3 → p13.2:).ish add(12)(8M16−,wcp12 +,TEL-ETV6 ++,RM2196 +).
Finally, all cases were investigated using a probe AHT specific for the TTAGGG telomere repeat sequence. All of the abnormal chromosomes described above showed signals with this probe at the terminus of the abnormal chromosome arm.
DISCUSSION
The high-resolution banding now attained by routine G-banding procedures is resulting in the detection of increasingly subtle abnormalities and rearrangements, and in the case of apparently terminal deletions, the subsequent use of subtelomere-specific probes and multisubtelomere fluorescence in situ hybridization (multisubtelomere FISH) allows the classification of such deletions as follows: (1) “true” terminal deletions where a region including the chromosome-specific subtelomeric region has been deleted; (2) interstitial deletions where a region proximal to, but not including, the subtelomeric sequence has been deleted; (3) interchromosomal (or intrachromosomal) rearrangements where the aberrant chromosome has lost a terminal segment including its subtelomeric region but where this has been replaced by a terminal segment, often submicroscopic, from another chromosome (or the same chromosome) and including the subtelomeric region in that segment. Other more complex rearrangements may also be elucidated.
Thirteen of the 16 (81%) cases with apparently terminal deletions described in this study were shown to be “true” terminal deletions, that is, they were missing the specific subtelomeric region of the deleted chromosome arm and did not have any other chromosome subtelomeric region present at the terminus. In addition, all 13 had a region of TTAGGG telomere repeats at their deleted termini. The presence of these telomere repeats can be explained by three different means: (a) they may have been synthesized in situ at the deleted chromosome ends by the enzyme telomerase, a mechanism known as “telomere healing” [Varley et al., 2000]; low levels of telomerase activity have been reported in normal human lymphocytes [Fan et al., 2000], and telomerase is thought to heal and stabilize broken chromosomes in the human germline. Alternatively, a telomerase-independent mechanism for telomere maintenance may have been responsible, such as recombination with a telomere from another chromosome and use of this as a template for telomere extension, using DNA polymerase and the recombination proteins Rad51p and Rad52p [Dunham et al., 2000]. (b) Existing telomere repeat sequences may have been “captured” from another non-homologous chromosome by recombination between homologous sequences [Meltzer et al., 1993; Flint et al., 1996]. (c) The original telomere repeat sequences may have been distal to the deleted segment, in which case the deletion would have been, strictly speaking, an interstitial one.
The abnormal chromosomes present in the other three cases were shown to be derivative chromosomes containing at least the subtelomeric region from a donor chromosome. The derivative chromosome in SW resulted from a semi-cryptic rearrangement between the long arm of chromosome 1 and the long arm of chromosome 6, the final karyotype being 46,XX,der(1)t(1;6)(q44;q27)de novo.ish der(1)(CEB108+,D1S3739−,D6S2522+) (see Fig. 1).
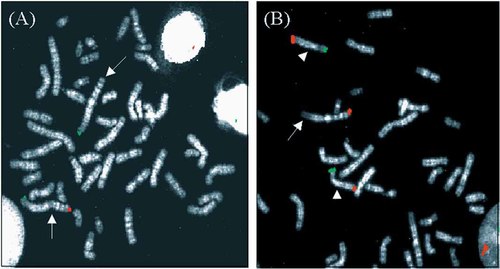
Multisubtelomere FISH on case SW. A: Probe D1S3739 (red) specific for the chromosome 1 long arm subtelomeric region, hybridized normally to one homolog of chromosome 1 but did not hybridize to the other (arrows). Probe CEB108+ (green) specific for the chromosome 1 short arm hybridized normally to both homologs. No hybridization was seen elsewhere. B: Probe D6S2522 (red) specific for the chromosome 6 long arm subtelomeric region, hybridized as expected to both homologs of chromosome 6 (arrowhead) but also hybridized to one homolog of chromosome 1 (arrow) at the distal end of the long arm. Probe 62I11 (green) specific for the chromosome 6 short arm subtelomeric region, hybridized normally to both homologs of chromosome 6. All other subtelomere probes hybridized normally.
Terminal deletion of chromosome 1q has been extensively reported in the literature [Meinecke and Vogtel, 1987; Garani et al., 1998; Ioan et al., 1992]. Many of these published cases involve larger regions than that seen in SW, often 1q42 → 1qter or 1q43 → 1qter and define a clinically recognizable syndrome. Most are de novo, the probands being born to young parents, as was the case with SW. Males and females are equally affected, the features including microcephaly, moderate to severe mental retardation, pre- and post-natal growth retardation, generalized seizures with reduced life expectancy, brachycephaly, down-slanting palpebral fissures, down-turned corners of the mouth, low-set ears, high-arched palate, short neck, and genital anomalies in males. Interestingly, although the deleted region of 1q in SW is smaller than that described in these publications, the phenotype is more severe with severe structural brain anomalies and skull asymmetry. It is possible that some of her phenotype is attributable to the extra copy of the terminal region of chromosome 6q, which has been associated with tetralogy of Fallot, cerebral malformations, thin lips, and an extremely short-webbed neck [Schmid et al., 1979; Turleau and de Grouchy, 1981; Franchino et al., 1987].
Case LS was shown by multisubtelomere FISH to have a semi-cryptic rearrangement between the long arm of chromosome 2 and the long arm of chromosome 5, with the karyotype 46,XX,der(2)t(2;5)(q37.2;q35.3).ish der(2)(D2S2983+,D2S2986−,D5S2097+). Terminal deletions of 2q37 → qter have been associated with developmental delay, hypotonia, psychomotor retardation, and brachydactyly [Lin et al., 1992; Conrad et al., 1995; Wilson et al., 1995]. These characteristics seem to encompass the phenotype of this case well, and there may therefore be no contribution from the very small trisomic segment of chromosome 5. Trisomy for the region 5q35 → 5qter has been described in conjunction with monosomy for other chromosome segments [Barber et al., 1996; Groen et al., 1998] and the associated phenotypes have included hypotonia, developmental delay, and brachydactyly.
ME was the third case shown to have a semi-cryptic rearrangement using multisubtelomere FISH, the chromosome abnormality being the result of a rearrangement between the short arm of chromosome 4 and the long arm of chromosome 11. The final karyotype was 46,XX,der(11)t(4;11)(p16.3;q24.1)de novo.ish der(11)(D11S2071+,D11S4974−,RM2072−,WHS−, GS10K2+). Deletions of this region of the long arm of chromosome 11 are associated with the characteristics of Jacobsen syndrome, which has been described in over 90 individuals [Jacobsen et al., 1973; Leegte et al., 1999]. ME was referred as a newborn with cardiac and brain anomalies, absent left kidney and dysmorphic features, and with a request to exclude trisomy 18. However in retrospect, it was felt that she did have some features consistent with Jacobsen syndrome those being the cardiac anomalies, flat nasal bridge, bossing forehead, low set ears, small mouth, structural eye abnormalities, and contracture of the fingers. Interestingly, Case 1 reported by Leegte et al. [1999] had a terminal deletion of the same portion of chromosome 11 (q24 → qter) and agenesis of the left kidney, although this has not been reported elsewhere. Trisomy for a larger portion of the short arm of chromosome 4 (4p15 → 4pter) has been associated with a bossed forehead and mental retardation [Clark et al., 1982] in a patient with duplication of this region due to adjacent-1 segregation of a balanced 4;14 translocation in the mother; the associated monosomy is thus only for the chromosome 14 short arm and the imbalance can be thought of in terms of pure partial 4p duplication.
In case CS, the abnormal chromosome 12p was found to have no chromosome-specific subtelomeric region from any chromosome, although the telomeric repeat sequence (TTAGGG) was present. This chromosome was shown by further FISH investigations using a more proximal short arm probe and chromosome 12-specific whole chromosome paint, to be composed entirely of chromosome 12 material and to have an inverted duplication of a small region of 12p and deletion of the 12p subtelomeric region. It is likely that this anomaly has arisen as a result of crossing over (either between sister or non-sister chromatids) following illegitimate pairing of direct and inverted repeat motifs. Such an event inevitably leads to an inverted duplication (with an intervening non-inverted segment), and deletion of the sequences distal to the most distal repeat motif, as described by Jenderny et al. [1998] and Bonaglia et al. [2000]. We are not aware of any previous description of such an event involving chromosome 12, although a direct duplication of this region has been described [Zelante et al., 1994]. The case reported here thus suggests that direct and inverted repeats may occur in this region of chromosome 12 as population variants, as has been suggested for chromosome 8 [Floridia et al., 1996].
Multisubtelomere testing of terminal anomalies, in combination with locus-specific probes and also potentially with terminal breakpoint analysis [Varley et al., 2000], is likely to uncover further cases involving complex rearrangements and should lead to a greater understanding of the mechanisms by which these abnormalities arise. In addition, more accurate ascertainment of genomic imbalance (as in 19% of the cases reported here) will provide valuable predictive phenotypic information in cases of semi-cryptic rearrangements.
Acknowledgements
The authors acknowledge the contribution of the other staff in the Cytogenetics Department.




