Integration of transcriptional and mutational data simplifies the stratification of peripheral T-cell lymphoma
Funding information Associazione Italiana Contro le Leucemie - Linfomi e Mieloma; Associazione Italiana per la Ricerca sul Cancro, Grant/Award Numbers: #10136, #14346, #16722, #17658
Abstract
The histological diagnosis of peripheral T-cell lymphoma (PTCL) can represent a challenge, particularly in the case of closely related entities such as angioimmunoblastic T-lymphoma (AITL), PTCL-not otherwise specified (PTCL-NOS), and ALK-negative anaplastic large-cell lymphoma (ALCL). Although gene expression profiling and next generations sequencing have been proven to define specific features recurrently associated with distinct entities, genomic-based stratifications have not yet led to definitive diagnostic criteria and/or entered into the routine clinical practice. Herein, to improve the current molecular classification between AITL and PTCL-NOS, we analyzed the transcriptional profiles from 503 PTCLs stratified according to their molecular configuration and integrated them with genomic data of recurrently mutated genes (RHOA G17V, TET2, IDH2 R172, and DNMT3A) in 53 cases (39 AITLs and 14 PTCL-NOSs) included in the series. Our analysis unraveled that the mutational status of RHOA G17V, TET2, and DNMT3A poorly correlated, individually, with peculiar transcriptional fingerprints. Conversely, in IDH2 R172 samples a strong transcriptional signature was identified that could act as a surrogate for mutational status. The integrated analysis of clinical, mutational, and molecular data led to a simplified 19-gene signature that retains high accuracy in differentiating the main nodal PTCL entities. The expression levels of those genes were confirmed in an independent cohort profiled by RNA-sequencing.
1 INTRODUCTION
Peripheral T-cell lymphomas (PTCL) represent a heterogeneous group of nodal and extra-nodal mature T-cell Non-Hodgkin lymphomas accounting for approximately 10%-15% of all lymphoma in the Western countries.1 Histological diagnosis of the various PTCL subtypes can still represent a challenge and difficulties occur in particular for those samples with borderline features between angioimmunoblastic T-cell lymphoma (AITL), follicular T-cell lymphoma, and PTCL-not otherwise specified (PTCL-NOS).1, 2 Previous studies have shown that these entities might bear distinct transcriptional and mutational profiles.3-8 Gene expression profiling has the potential to represent the gold standard for classification, but its clinical use is still limited due to technical limits and to the absence of a manageable and practical short consensus gene signature. Recent advances in next-generation sequencing (NGS) allowed the discovery of recurrently mutated genes (RHOA, TET2, DNMT3A) in approximately 60%-70% of AITL and in 20%-30% of PTCL-NOS, changing in part this landscape.6, 9-12 Notably, 20%-30% of AITL cases can carry hotspot IDH2 R172 mutations that are virtually absent in PTCL-NOS.9 Nevertheless, these findings have not yet significantly impacted diagnosis in daily clinical practice, which largely relies on morphological and immunophenotypic features of tumor cells.1 Moreover, albeit some mutations appear to be linked to distinct transcriptional signature(s),6 the full potential of an integrated genotypic-transcriptomic analysis has not been thoroughly tested in PTCLs. Herein, we collected a large gene expression profiling data set of PTCLs, and performed an integrative analysis with available mutational data to improve our understanding of the underlying structure of sample clusters, with potential implications for disease classification particularly at the interface between AITL and PTCL-NOS lymphomas.
2 METHODS
2.1 Data set
We analyzed 503 PTCL samples, univocally acquired from 8 studies (GSE6338, GSE14879, GSE19067, GSE19069, GSE58445, and GSE65823 at http://www.ncbi.nlm.nih.gov/geo/; ETABM702 and ETABM783 at https://www.ebi.ac.uk/arrayexpress, Supporting Information Figure S1). Normalized data were extracted using RMA procedure and the annotation available at http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/21.0.0/entrezg.asp. A batch-effect correction was applied using ComBat function in sva package for R software. The whole data set with all available clinical and genomic information acquired was uploaded to https://github.com/emacgene/PTCL.
2.2 Transcriptional and statistical analysis
The statistical models that allow measuring the association between mutations and gene expression was firstly described in Gerstung et al.13 and here adapted to 39 AITLs and 14 PTCL-NOSs for whom mutational data for IDH2 R172, DNMT3A, TET2, and RHOA G17V were available.6
ConsensusClusterPlus package for R14 was used to determine the significance and robustness of natural grouping of patients based on selected transcriptional data, using Ward and Euclidean as linkage and distance metrics, respectively.
CIBERSORT analysis was performed as previously described, using standard procedure and LM22 signature.15 The CIBERSORT different contribution for each signature was then tested by pairwise.wilcox.test R function. Benjamini-Hochberg correction was used for multiple testing adjustment. The pathway enrichment analysis was performed using different modalities. The tmod R package was used on limma-derived signatures to decipher whether clusters deregulate blood cell-associated transcriptional modules described by Chaussabel et al.16 and by Li et al.,17 according to the procedure described by Weiner et al.18 The full analysis process written in R is provided in Supplementary Data at https://github.com/emacgene/PTCL.
Data from 34 previously published RNAseq samples have been imported upon obtainment of accession to dbGap data set #phs000689.v1.p1.9 RNAseq mapping and read counts were processed using iRAP pipeline,19 and the RNAseq raw expression data were normalized as previously described.20
3 RESULTS
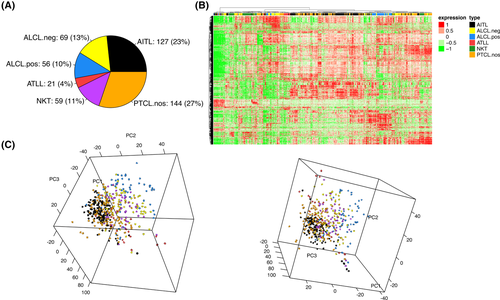
According to the most recent updates in the T-cell lymphoma classification1 and the gene-expression based classification criteria as in Iqbal et al.,21 we generated the whole transcriptional profile of a data set including 127 AITL, 144 PTCL-NOS, 56 ALK+ (anaplastic large-cell lymphoma) ALCL, 96 ALK− ALCL, 21 Adult T-Cell Lymphoma (ATLL), 59 NK/T-cell lymphomas (Figure 1A, from here on named as “molecular classification”). Both unsupervised hierarchical clustering and principal component analysis on the most variable genes (exceeding the mean an average 2-fold across the data set) showed that the known entities, such as ALK+ ALCLs and ATLL were associated with markedly distinct signatures; notably, the transcriptional portrait of AITL and PTCL-NOS displayed a considerable overlap (Figure 1B,C). For completeness, the stability of the identified clusters was tested to unravel the most relevant overlapping and to describe the phenotypes characterized by overall uniform transcriptional pattern; the whole confusion matrix was reported in Supplementary Data. Overall, our meta-analysis of the largest gene-expression profiling data set tested to date showed that the consensus between transcriptomic analysis and histology is still imperfect, opening the field for the search of additional features that could improve diagnostic accuracy.
3.1 Definition of gene signature associated with molecular classification and mutational status
To search for additional features that could better define the AITL and PTCL-NOS entities, we investigated whether recurrent mutations or clinical features might correlate with a specific transcriptional pattern that could be used for stratification purposes. To this aim, we adapted a recently published analysis13 to a set of 39 AITL and 14 PTCL-NOS cases for which mutational data for IDH2 R172, DNMT3A, TET2, and RHOA G17V were available.6 This analysis allowed the creation of a linear model that associated gene-by-gene expression to putative predictor variables, namely the molecular classification histotype, mutations, age, and gender of each patient. We identified 221 modulated genes across the training data set at false discovery rate (FDR) <1% (Figure 1A), among which 30 of them emerged as significantly associated with 1 distinct variable (Figure 2B). Gender selectively impacted 14 genes located on the X or Y chromosomes, which were discarded from further analysis. Histotype and IDH2 R172 mutations specifically impacted the expression of 13 and 3 genes, respectively, while notably the mutational status of RHOA G17V, TET2, and DNMT3A (Figure 2B) were not associated with any distinct gene expression change. The 3 genes associated with IDH2 R172 mutation were ID2, NETO2, and SLC5A3. Three out of 13 histotype-associated genes were also reported in a previous large gene expression signature that discriminates PTCL-NOS and AITL (ROBO1, ARHGEF10, and EFNB2).3 Based on these findings, we wondered whether a minimal signature including these 16 genes might effectively stratify AITL and PTCL-NOS cases. A leave-one-out cross validation (LOOCV) procedure run using linear discriminant analysis22 supported the robustness of this 16-gene model, indicating overall 86.4% accuracy (sensitivity 86.9%; specificity 85.9%) in discriminating AITL from PTCL-NOS.
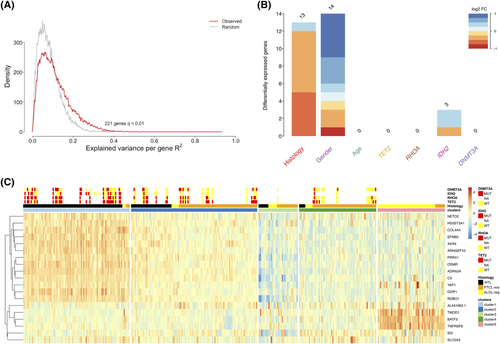
Prompted by the evidence that this small 16-gene signature robustly stratified these 2 entities, we hypothesized that combining this signature with the previously described 3-gene ALK− ALCL signature22 may lead to a 19-gene model able to improve the stratification of all major PTCL entities (ALK− ALCL, PTCL-NOS and AITL). Again, a LOOCV procedure indicated that the 19-gene model was robust (overall accuracy 80.1%).
3.2 Assessment of the 19-gene signature expression level in an independent cohort
To test the reproducibility of the signature, we analyzed the expression levels of the genes for which data were available in 30 PTCL-NOSs cases (11 AITL, 11 PTCL-NOS, 8 ALK− ALCL) from a previously published RNAseq data set.9 To this aim, we first classified the samples according to the described molecular classification using a LOOCV procedure and the published gene signature3 that has been used to test build the expression/mutation model (Supplementary data). Of the 19 genes included in our model, 17 were mapped and detected in the 30 samples, overall retaining significant difference in the group distribution (Supporting Information Figure S2).3, 22
3.3 Patients' stratification according to the 19-gene signature
To further investigate the significance and robustness of natural grouping of patients based on selected 19-gene transcriptional data, we applied the ConsensusClusterPlus package to our original cohort, using Ward and Euclidean as linkage and distance metrics, respectively. This analysis led to the recognition of a minimum of 5 distinct subsets at the highest significance (Figure 2C), whose features are summarized in Table 1. The first group (C-1; 87 cases) mostly included AITL samples (93%; 81/87) enriched for IDH2 R172 (16/36; 44%) and RHOA G17V (23/29; 79%) mutations. The second cluster (C-2; 103 samples), where the 19-gene expression pattern resembled that in C-1 but at lower levels, included samples annotated either as PTCL-NOS (64/103; 62%) or AITL (33/103; 32%). Interestingly, in the C-2 group, despite the IDH2 R172 mutation was detected in only 1 out of 23 cases where the mutational status was known (P = .001 if compared to C-1), a very high prevalence of RHOA G17V mutations was observed (15/29; 52%). RHOA G17V mutation occurrence was, moreover, significantly higher than what observed in clusters 3 and 4 (P < .005). Despite their high prevalence, RHOA G17V mutations had a lower impact on transcription (as evidenced by Figure 2B), likely because they were prevalently detected at sub-clonal level in a significant fraction of these cases, and this may explain their specific lower impact on transcription (as evidenced by Figure 2B). The third cluster (C-3; 32 samples) included patients who showed the lowest 19-gene expression levels overall, and an equal admixture of the 3 main PTCL entities. Group 4 (C-4; n = 63) included mainly PTCL-NOS (52/63; 82%), showing low prevalence of RHOA G17V and TET2 mutations compared either to C-1 (P < .0001 and P = .0002, respectively) or C-2 (P = .005 and .03, respectively). In the last cluster (C-5; n = 55) ALK− ALCL cases (47/69; 68%) were over-represented, confirming the strong association between these lymphomas and the previously published expression pattern of TNFRSF8, BATF3, and TMOD1.22
| Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 | Cluster 5 | |
|---|---|---|---|---|---|
| Histology | |||||
| AITL | 81 (93%) | 33 (32%) | 8 (25%) | 5 (8%) | 0 |
| ALCL ALK-neg | 3 (3.5%) | 6 (6%) | 7 (22%) | 6 (9.5%) | 47 (88%) |
| PTCL-NOS | 3 (3.5%) | 64 (62%) | 17 (53%) | 52 (82.5%) | 8 (12%) |
| Gender | |||||
| Female | 26 (37%) | 31 (41%) | 4 (33%) | 20 (41%) | 8 (26%) |
| Male | 45 (63%) | 43 (59%) | 8 (67%) | 28 (59%) | 22 (74%) |
| Age (Median) | 64 (33-87) | 62 (19-88) | 60 (35-97) | 62 (21-87) | 59 (31-80) |
| TET2 | |||||
| MUT | 26 (84%) | 19 (65%) | 0 (0%) | 6 (32%) | 0 |
| WT | 5 (16%) | 10 (35%) | 1 (100%) | 13 (68%) | 0 |
| DNMT3A | |||||
| MUT | 13 (42%) | 9 (31%) | 0 | 3 (16%) | 0 |
| WT | 18 (58%) | 20 (69%) | 1 (100%) | 16 (84%) | 0 |
| RHOA | |||||
| MUT | 23 (79%) | 15 (51%) | 0 | 2 (10.5%) | 0 |
| WT | 6 (21%) | 14 (49%) | 1 (100%) | 17 (89.5%) | 0 |
| IDH2 | |||||
| MUT | 16 (45%) | 1 (4%) | 0 | 1 (33%) | 0 |
| WT | 20 (55%) | 22 (96%) | 0 | 2 (67%) | 0 |
3.4 Functional annotation, gene-expression-based estimation of microenvironment composition, and clinical relevance of 19-gene associated groups
To decipher whether these clusters might be characterized by common transcriptional behavior, we first investigated if they showed pathway enrichments. To this aim, tmod R package was used on limma-derived signatures to query whether clusters deregulated specific transcriptional modules associated with distinct blood cells. This analysis suggested significant enrichment of B-cell-related pathways in C-1 and C-2, whereas C-5 showed enrichment for stimulated CD4+ T cells associated pathways, and significantly lower involvement of T-cell differentiation and T-cell activation pathways (Supporting Information Figure S3). To understand whether this might be the consequence of a somehow unbalanced microenvironment composition, we then applied CIBERSORT,23 an analytical tool developed to provide an estimation of the abundances of specific cell types in a mixed cell population based on gene expression data. CIBERSORT analysis was performed as previously described, using standard procedure and the LM22 signature (Supporting Information Figure S4).23 Notably, we observed a prevalence of pathways associated with plasma cells in C-1 and C-2, in agreement with the notion that AITL are often enriched in B-cells/plasma cells.3 Also of interest, CIBERSORT analysis evidenced a slight prevalence of activated NK cell-like profiles in the C-3 cluster, overall suggesting that this cluster might be considered a spurious entity due to partial contamination of NK-associated signatures; and higher presence of activated memory T-CD4 lymphocytes in the ALK− ALCL cluster (C-5), supporting previous pieces of evidence that CD4+ expression characterizes almost all the anaplastic lymphomas,24 and is not exclusively dependent on the ectopic expression of the ALK protein.
Finally, we tested whether the 5 clusters were associated with different prognosis in the 239 patients for whom outcome data were available. No significant global differences in prognosis could be appreciated; however, a marginally poorer prognosis was identified in C-3 patients (P = .06, Supplementary Data and Supporting Information Figure S5).
4 DISCUSSION
Gene expression has emerged as one of the most robust and reliable approaches to differentiate human lymphomas. This has also been the case of T-cell lymphomas, where gene expression-based strategies have allowed the distinction of closely related entities such as AITL and PTCL-NOS.3, 21, 22, 25-28 However, gene expression profiling from formalin-fixed paraffin-embedded samples has known technical artifacts29 and analysis is not standardized. Thus, the molecular/expression stratification of PTCL has not entered yet into routine clinical practice. Recently, novel technologies (eg, nanoString) have provided reproducible and feasible quantification of specific transcripts from FFPE samples, particularly for diffuse large B-cell lymphoma.30-32 However, this approach has not yet been tested in PTCL, likely because a short and robust list of differentially expressed genes has not emerged yet.
To bridge this gap, we performed a meta-analysis of a large PTCL series, assessing the molecular profile of the main PTCL subgroups and defining a manageable list of significant differentially expressed genes. By integrating transcriptional and NGS data, we were able to discover the transcriptional impact of the recurrent mutations in AITLs and PTCL-NOSs. Interestingly, TET2, DNMT3A, and RHOA G17V mutations were not associated with a distinct gene expression signature. In particular, RHOA G17V mutations were detected at sub-clonal level in a significant fraction of these cases (43%) and this may explain its low impact on transcription.6 On the other hand, TET2 and DNMT3A mutations are likely clonal mutations arising from hematopoietic stem cells involved in clonal hemopoiesis;33-35 therefore, these mutations could induce gene expression signatures shared by other PTCL subtypes and masked by the molecular classification, subsequently limiting its extraction through our statistical process. Conversely, in line with a previous report,6 the IDH2 R172 mutation significantly correlated with a distinct expression signature independently from the molecular subgroup. Through the integration pathological data, transcriptional, and NGS analyses, we then defined a short list of 19 genes whose differential expression divided the samples into 5 clusters, strongly associated with distinct PTCL entities. Specifically, C-1 and C-2 were characterized by most of the AITL hallmarks. In addition, despite their limited gene expression impact, the great majority of RHOA G17V and TET2 mutations were grouped together, confirming their potential utility in the diagnostic process of these lymphomas. A significant fraction of PTCL-NOSs were included in these 2 “AITL clusters.” Most of these cases showed high prevalence of RHOA G17V and TET2 mutations and expressed some distinct, and in part previously reported, AITL-associated genes. Unfortunately, missing paired molecular/genomic data in a significant portion of the data set prevented the possibility to define a robust and definitive molecular classifier able to dissect this “gray zone” of PTCL classification.
Nonetheless, 6 of the 16-gene signature were previously described as T follicular helper (TFH)-phenotype related.8 Unfortunately, no cases could be a priori classified as nodal PTCL-NOS with (TFH) phenotype, since WHO has introduced this category after the original studies were performed1 and phenotypic data were not available. However, these 6 genes were significantly overexpressed in C-1 and C-2, in line with the recent WHO classification where PTCL TFH and AITL are grouped under an unique umbrella category.
Finally, based both on the present and recently published3, 21, 36 data, PTCL-NOS cases in cluster C-4 likely may represent a distinct biologically entity. In line with previous findings,4 cases within this cluster had a distinct genotypic and transcriptional pattern, mostly characterized by significant enrichment in downregulated genes.
Taken together, our analyses based on a small 19-gene model offered a more insightful perspective of the dissection of the complex nodal PTCL entities. We suggest that, in the future, the integration of transcriptional and genotypic data may improve the identification of clinic-pathological entities, contributing to the current diagnostic approach of PTCLs and laying the basis for effective treatments through the identification of specifically and recurrently dysregulated pathways.
ACKNOWLEDGMENT
NB is funded by A.I.R.C. (Associazione Italiana per la Ricerca sul Cancro) through a MFAG #17658. FM is supported by A.I.L. (Associazione Italiana Contro le Leucemie-Linfomi e Mieloma ONLUS). AN is funded by A.I.R.C. Investigator grants #16722 and #10136. PC is funded by A.I.R.C. Investigator grant #14346.
CONFLICT OF INTEREST
No conflict of interests to declare.
AUTHOR CONTRIBUTIONS
Study design: Maura, Agnelli, Corradini
Data collection: Maura, Agnelli, Corradini, Chan, Heavican, Dodero, Carniti, Pellegrinelli, Pruneri, Butler, Chiappella, Di Rocco, Zinzani, Zaja, Piva, Iqbal, Palomero
Data analysis: Maura, Agnelli, Corradini, Leongamornlert, Wang, Campbell, Bhosle
Manuscript writing: Maura, Agnelli, Corradini
Critically revised the manuscript: Bolli, Neri, Heavican




