Automated screening for myelodysplastic syndromes through analysis of complete blood count and cell population data parameters
Conflicts of interest: GJG and TN participated in scientific advisory board meetings organized by Beckman Coulter and visited an international congress for which travel and accommodation expenses were reimbursed by Beckman Coulter. DS has been employed by Beckman Coulter as an independent consultant. GJG and GW have received a speaker fee from Beckman Coulter.
Abstract
The diagnosis of myelodysplastic syndromes (MDS) requires a high clinical index of suspicion to prompt bone marrow studies as well as subjective assessment of dysplastic morphology. We sought to determine if data collected by automated hematology analyzers during complete blood count (CBC) analysis might help to identify MDS in a routine clinical setting. We collected CBC parameters (including those for research use only and cell population data) and demographic information in a large (>5,000), unselected sequential cohort of outpatients. The cohort was divided into independent training and test groups to develop and validate a random forest classifier that identifies MDS. The classifier effectively identified MDS and had a receiver operating characteristic area under the curve (AUC) of 0.942. Platelet distribution width and the standard deviation of red blood cell distribution width were the most discriminating variables within the classifier. Additionally, a similar classifier was validated with an additional, independent set of >200 patients from a second institution with an AUC of 0.93. This retrospective study demonstrates the feasibility of identifying MDS in an unselected outpatient population using data routinely collected during CBC analysis with a classifier that has been validated using two independent data sets from different institutions. Am. J. Hematol. 89:369–374, 2014. © 2013 Wiley Periodicals, Inc.
Introduction
Myelodysplastic syndromes (MDS) are a group of clonal hematopoietic diseases, primarily of older individuals, that are characterized biologically by ineffective hematopoiesis, morphologically by dysplasia in at least one hematopoietic cell line, hematologically by anemia, leukopenia, and thrombocytopenia, and clinically by symptoms related to these cytopenias 1. With the aging of the population and sometimes-subtle presentation, there exists a need to optimize diagnostic tools 2, 3. Diagnosing MDS essentially always includes a bone marrow aspirate and biopsy to assess cellularity, dysplasia, and clonal cytogenetic abnormalities, with molecular testing poised to assume an increasingly important role. However, the initial assessment of dysplasia is sometimes subjective, particularly in the early phases of the disease, and cytogenetic changes are seen in only ∼50% of de novo cases 4. Hence, there is a need for more robust diagnostic tools. While characteristic morphologic changes can be seen in peripheral blood leukocytes, this assessment is similarly limited by subjective and labor-intensive manual analysis. Consequently, only a small fraction of peripheral blood samples receive a manual review. The development of an automated, minimally invasive, and objective assay to screen for MDS on peripheral blood samples could lead to robust prediction of MDS in patients undergoing routine laboratory analysis.
Automated hematology analyzers utilize various analytical techniques to quantitate peripheral blood cell types and classify leukocytes. The Beckman Coulter LH 750 and LH780 automated hematology analyzers generate complete blood count (CBC) and leukocyte differential count data based on three physical parameters: cell volume, conductivity, and light scatter (known collectively as cell population data [CPD]). Following quantitation and lysis of red blood cells, the volume of leukocytes is measured by direct current impedance, leukocyte conductivity by radio frequency opacity measurements, and cytoplasmic granularity by light scatter. This information is used to generate an automated differential count. Identification of major deviations in the abundance of cell types or identification of unexpected cell populations (e.g., blasts) is flagged and, in many laboratories, triggers a manual microscopic review of the associated blood smear.
In addition to identifying peripheral leukocyte subsets, automated hematology analyzers also generate data in identifiable patterns that correspond to numerous pathologic conditions. Algorithms that identify patients with bacterial infections 5-13, viral infections 14, malaria 15, asthma 16, 17, myocardial infarction 18, postprandial leukocyte activation 19, and chronic lymphoproliferative disorders 14, 20, 21 have been described on numerous commercially available hematology analyzers. These algorithms routinely generate data to predict whether patients have the condition in question. Theoretically, this can expedite diagnosis and therapy, while ideally minimizing additional manual review of peripheral smears or other testing. Clinical adoption of these algorithms has been variable.
The diagnosis of MDS through detection of peripheral blood abnormalities by an automated analyzer is a long-standing goal; the first attempts were described in 1985 22, 23. Our understanding of both MDS and automated analyzer technology has increased dramatically since then, but this goal remains elusive. Numerous studies have identified differences in hematology analyzer parameters between samples from normal patients and those with MDS 20, 24, 25, but data on utility of these algorithms in an unselected patient population is limited 26. We report herein a novel classification framework using CBC, CPD, and demographic parameters to identify patients with MDS in an unselected outpatient population.
Methods
All procedures were approved by the University of Pennsylvania Institutional Review Board. CBC data were collected retrospectively on 15,314 samples from 5,470 individual patients presenting to an outpatient medical clinic over a 6-month time period. The clinic serves hematology, oncology, and general medical outpatient populations. CBC analysis was performed using the Coulter LH 780 hematology analyzer; this instrument utilizes the Coulter principle, radio frequency conductivity, and optical detection to identify cell types. Data were collected on standardly reported CBC parameters, research use only (RUO) parameters, and CPD. Clinical and demographic information on patients was extracted from the electronic medical record.
Patients with a history of chemotherapeutic agent therapy were identified, and those patients who had received chemotherapeutic agents within three weeks prior to the CBC were excluded from analysis in order to minimize false positives potentially created by the transient effects of chemotherapy. The first sample for each patient during the study period was used for analysis; additional samples were excluded so as to not bias the database towards patients who had frequent CBCs. Patients with MDS were initially identified by screening the electronic medical record for any patient with an International Classification of Disease, 9th revision (ICD-9) code corresponding to MDS; clinical data from these patients were examined in detail, and only those patients with a confirmed hematopathologic diagnosis of MDS were included in the MDS study group.
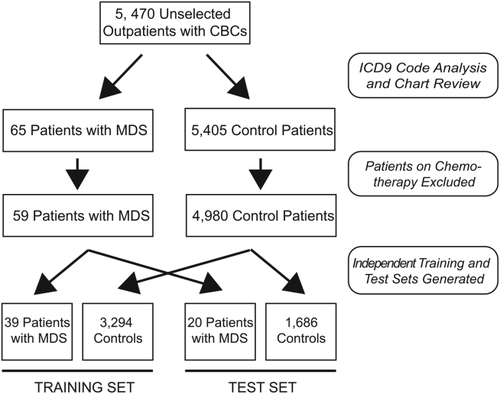
Patient CBCs were randomized and divided into independent, nonoverlapping training and test sets containing proportional numbers of patients with MDS and controls. CBCs with incomplete information (n = 5, all control patients) were removed from subsequent analysis following randomization and separation into training and test sets. The training set consisted of 39 CBCs from patients with MDS and 3,294 CBCs from control patients, and the test set consisted of 20 CBCs from patients with MDS and 1,686 CBCs from control patients. Selection of patient CBCs and separation into training and test sets is summarized in Fig. 1.
Data collected on the Coulter LH 780 instruments includes parameters that are typically reported clinically and unreported RUO and CPD parameters (dataset available as Supporting Information). Briefly, a random forest classifier for MDS status was trained on the training data set using the randomForest package version 4.6–7 in the R statistical program version 2.15.3 27, 28. R code is available as a Supporting Information. The random forest approach has been successfully applied in biomedical datasets with large sample size imbalances between affected and unaffected groups 29, and given the imbalance in our training set (39 MDS patients and 3,294 control patients) we used balanced random subsamples (39 cases, 39 controls) to generate 5,000 trees in the random forest. Following classifier generation, a separate test set (not utilized in classifier development) was evaluated. Classifier performance was evaluated via receiver operating characteristic (ROC) curve analysis and standard measures, and calculation of the area under the curve (AUC) was performed using the ROCR package version 1.0–4 in R 30.
An independent dataset collected on similar instruments (Beckman Coulter LH 750) at a different institution (Sint Franciscus Gasthuis, Rotterdam, The Netherlands) consisted of 239 patients. Eight of these patients had a diagnosis of MDS and the remaining 231 were controls consisting of healthy volunteers or patients visiting the outpatient clinic for treatment of diabetes and primary or secondary prevention of cardiovascular disease. This dataset was used as an additional, separate test set for the random forest classifier. Data collected from the Beckman Coulter LH 750 instruments does not include the parameters standard deviation of red cell distribution width (RDW-SD), uncorrected white blood cell count (UWBC), and microcytic anemia factor (MAF), a proprietary measurement created by combining hemoglobin and MCV. These variables were omitted from the classifier during analysis of this dataset (dataset available as a Supporting Information).
During routine clinical automated hematology analysis, samples with marked abnormalities in any measured or calculated parameter were identified by standard flagging algorithms. Flagged samples were then manually screened by a hematology technologist to confirm the abnormality, perform a manual differential, or identify blasts. A subset of these samples was also reviewed by a hematopathologist.
Concordance between the random forest classifier results and bone marrow biopsy results was also investigated. Patients in the test set were included if they met all three of the following criteria: (1) patients who had both a CBC and bone marrow aspirate and biopsy during the study period; (2) patients who did not have a prior myeloid malignancy or acute leukemia, or patients who had MDS but had undergone an allogeneic stem cell transplant and did not have evidence of recurrence; (3) patients who had a clinical history accompanying the bone marrow aspirate and biopsy of “rule out MDS” or cytopenias. A total of 359 patients had both CBCs and bone marrow biopsies during the study period, and 6 patients in the test set met all the aforementioned criteria. Patients in the training set were excluded from this analysis.
Results
Patient selection
To develop an algorithm that identifies patients with MDS, CBC data were retrospectively collected from 15,314 sequential samples from 5,470 patients over a 6 month period. CBCs from 5,039 patients met criteria described above, of whom 59 with MDS were identified (1.2% of analyzed patients); the remaining 4,980 served as controls (Fig. 1).
Univariate analysis of differences between MDS and control patients
Univariate analysis of demographic and CBC parameters identified numerous key differences between patients with MDS and the control group (Table 1). On average, patients with MDS were ∼9 years older than control patients. Patients with MDS also had significantly decreased hematocrit, white blood cell count, and platelet counts as compared to controls. The mean corpuscular volume (MCV) was elevated (but still within the normal range) in patients with MDS, and red blood cell distribution width (RDW), the standard deviation of the RDW (RDW-SW), and platelet distribution width (PDW) were all increased in patients with MDS. Automated differential blood counts demonstrated significantly decreased percentage neutrophils and correspondingly increased percentage lymphocytes in patients with MDS.
| Non-MDS | MDS | ||||
|---|---|---|---|---|---|
| Average | SD | Average | SD | t-test | |
| Age (y) | 59.6 | 13.9 | 69.0 | 11.7 | 4.5 × 10−8 |
| WBC (× 103/ml) | 7.1 | 5.9 | 4.4 | 3.2 | 0.008 |
| Hgb (g/dl) | 12.8 | 1.9 | 10.3 | 1.8 | 5.4 × 10−27 |
| Hct (%) | 37.4 | 5.3 | 30.4 | 5.3 | 1.4 × 10−26 |
| Plt (× 103/ml) | 225 | 100 | 173 | 224 | 3.7 × 10−5 |
| MCV (fl) | 92.4 | 7.0 | 97.0 | 9.9 | 1.7 × 10−7 |
| MCH (pg/cell) | 31.6 | 2.8 | 32.9 | 3.7 | 8.9 × 10−5 |
| MCHC (g/dl) | 34.1 | 0.9 | 33.9 | 1.1 | 0.10 |
| RDW (%) | 15.1 | 2.6 | 18.9 | 4.5 | 2.8 × 10−31 |
| RDW-SD | 48.1 | 8.4 | 63.4 | 14.8 | 2.4 × 10−48 |
| PDW (%) | 17.1 | 0.7 | 18 | 1.0 | 6.8 × 10−26 |
| % Ne | 63.1 | 14.5 | 51.5 | 20.4 | 2.5 × 10−10 |
| % Ly | 24.8 | 13.4 | 36.1 | 19.0 | 1.9 × 10−11 |
| % Mo | 9.0 | 5.0 | 10.0 | 8.7 | 0.10 |
| % Eo | 2.5 | 2.9 | 1.8 | 1.7 | 0.03 |
| % Ba | 0.6 | 0.6 | 0.6 | 0.7 | 0.74 |
| Abs Ne (cells/ml) | 4.4 | 4.1 | 2.6 | 2.7 | 0.0007 |
| Abs Ly (cells/ml) | 1.9 | 6.1 | 1.2 | 0.5 | 0.35 |
| Abs Mo (cells/ml) | 0.6 | 1.2 | 0.4 | 0.7 | 0.33 |
| Abs Eo (cells/ml) | 0.2 | 0.3 | 0.1 | 0.1 | 0.05 |
| Abs Ba (cells/ml) | 0.04 | 0.1 | 0.02 | 0.1 | 0.36 |
- Patients with MDS have significantly decreased hemoglobin and hematocrit, WBC count, platelet count. MCV, MCH, RDW, RDW-SD, and PDW are all significantly elevated in patients with MDS. Patients with MDS also show a decrease in the percentage neutrophils and concordant increase in the percentage lymphocytes.
- CBC: complete blood count; MDS: myelodysplastic syndrome; SD: standard deviation; WBC: white blood cell count; Hgb: hemoglobin; Hct: hematocrit; Plt: platelet; MCV: mean corpuscular volume; MCH: mean corpuscular hemoglobin; MCHC: mean corpuscular hemoglobin concentration; RDW: red blood cell distribution width; RDW-SD: standard deviation of the red blood cell distribution width; PDW: platelet distribution width; % Ne: percentage neutrophils; % Ly: percentage lymphocytes; % Mo: percentage monocytes; % Eo: percentage eosinophils; % Ba: percentage basophils; Abs: absolute count.
Individual CPD parameters for neutrophils, monocytes, and lymphocytes showed trends toward differences between MDS and control populations that did not achieve significance following Bonferroni correction for multiple hypothesis testing (Table 2). Mean conductivity showed a trend toward lower values in the MDS population in all cell types examined, suggesting possible differences in internal composition of leukocytes in MDS. The standard deviation of neutrophil volume also showed a trend toward being increased in the MDS population.
| Non-MDS | MDS | ||||
|---|---|---|---|---|---|
| Average | SD | Average | SD | t-test | |
| Neutrophil | |||||
| Mean volume | 130.9 | 19.4 | 140.9 | 28.4 | 0.66 |
| Volume SD | 20.4 | 4.9 | 22.1 | 7.9 | 0.007 |
| Mean scatter | 138.1 | 19.0 | 133.1 | 24.7 | 0.03 |
| Scatter SD | 12.7 | 2.9 | 12.5 | 3.2 | 0.50 |
| Mean conductivity | 142.1 | 17.5 | 138.0 | 24.5 | 0.05 |
| Conductivity SD | 6.0 | 2.2 | 6.0 | 2.2 | 0.85 |
| Monocyte | |||||
| Mean volume | 165.2 | 21.6 | 166.9 | 31.5 | 0.53 |
| Volume SD | 18.1 | 17.7 | 18.2 | 18.0 | 0.77 |
| Mean scatter | 88.4 | 11.4 | 88.3 | 16.2 | 0.90 |
| Scatter SD | 10.0 | 1.7 | 9.9 | 2.1 | 0.32 |
| Mean conductivity | 119.1 | 14.7 | 115.8 | 20.5 | 0.07 |
| Conductivity SD | 4.3 | 1.1 | 4.3 | 1.2 | 0.95 |
| Lymphocyte | |||||
| Mean volume | 86.9 | 12.1 | 88.1 | 16.9 | 0.43 |
| Volume SD | 15.1 | 3.2 | 15.0 | 3.4 | 0.81 |
| Mean scatter | 69.5 | 10.1 | 70.0 | 14.3 | 0.68 |
| Scatter SD | 15.9 | 3.4 | 15.6 | 4.2 | 0.38 |
| Mean conductivity | 110.9 | 13.7 | 108.0 | 19.3 | 0.08 |
| Conductivity SD | 10.0 | 3.0 | 9.5 | 4.0 | 0.24 |
- CPD: cell population data; MDS: myelodysplastic syndrome; SD: standard deviation.
- Patients with MDS showed trends toward increased volume in neutrophils and lymphocytes, and a trend toward increased standard deviation of neutrophil volume. No parameter reached statistical significance following Bonferroni correction.
Random forest classifier detection of MDS
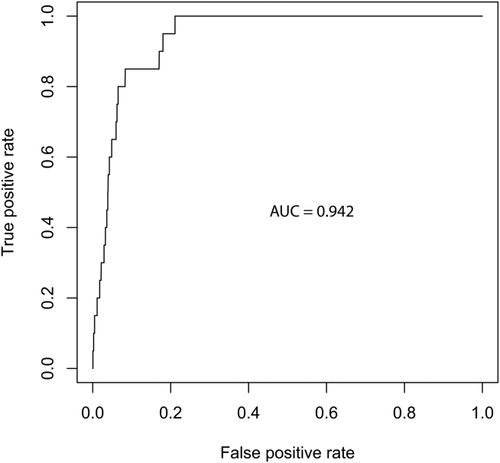
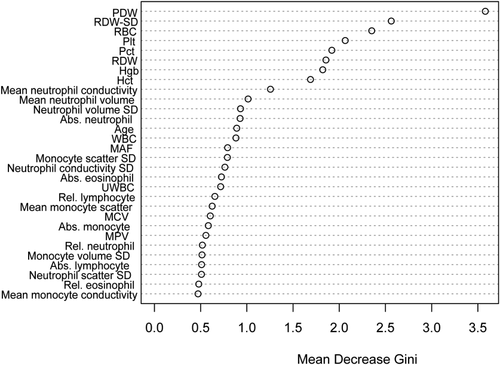
A random forest classifier was generated from the training set and applied to the independent test set. The classifier performed well in the test set, generating an ROC curve with an AUC of 0.942 (Fig. 2). Additional performance characteristics are listed in Table 3; the sensitivity of the classifier was 85.0% and specificity was 87.1%; therefore the false positive rate was 12.9% and the false negative rate was 15.0%. Importantly, given the relatively low prevalence of MDS in this unselected patient population, the negative predictive value of the algorithm was 99.8%. Conversely, the positive predictive value was 7.3%. Relative variable importance within the classifier, as determined by the mean decrease in the Gini coefficient, is shown in Fig. 3. The Gini index is a measure of the distribution of patient CBCs into MDS and non-MDS groups at a particular point in the random forest classifier 31. Variables with a high decrease in the mean Gini index are those which increase separation of the sample into MDS and non-MDS groups, i.e., variables with the greatest separating power. The RUO parameters platelet distribution width and the standard deviation of red cell distribution width are the most discriminating factors between MDS patients and controls. Mean neutrophil conductivity, mean neutrophil volume, and the standard distribution of neutrophil volume were the most important discriminating CPD parameters within the classifier.
| MDS random forest classifier | |||
|---|---|---|---|
| Diagnosis | Positive | Negative | |
| MDS | 17 | 3 | 85.0% Sensitivity |
| No MDS | 217 | 1,469 | 87.1% Specificity |
| 7.3% PPV | 99.8% NPV | ||
- MDS: myelodysplastic syndrome; PPV: positive predictive value; NPV: negative predictive value.
Feasibility of clinical implementation of the random forest classifier
The random forest classifier detects more patients with MDS than standard flagging algorithms and minimally increases the number of CBCs requiring additional manual screening. Whereas standard flagging algorithms only prompted subsequent peripheral blood smear review in 25% of patients with MDS, the random forest classifier identified 85% of patients with MDS in the independent test set. The standard flagging algorithm designated 8.0% of samples within the independent test set for subsequent peripheral blood smear analysis, versus 13.7% of samples designated by the random forest classifier. After accounting for those cases flagged by both approaches and scaling to reflect the entire study population and duration (15,314 CBCs over six months), clinical implementation of the MDS classifier for initial CBCs would result in an increase from 10.7 to 14.9 peripheral blood smear reviews per day.
In addition to screening for unsuspected MDS, we hypothesized that the random forest classifier could assist in the evaluation of patients with a clinical differential including MDS. A positive random forest classifier result could prompt an early bone marrow examination, or a negative result could support a more conservative approach and prevent an unnecessary invasive procedure. To assess performance in the most relevant clinical scenario, only those patients in the test set whose bone marrow biopsy came with the clinical information “rule out MDS” or a history of cytopenias were included in the analysis (n = 6). The random forest classifier accurately identified the three patients with MDS (including one with acute myeloid leukemia with myelodysplasia-related changes) and two patients without MDS. The remaining patient classification was a false positive; the classifier indicated MDS, whereas the bone marrow revealed primary myelofibrosis.
Additional independent cohort validation of random forest classifier
Having validated the performance of our classifier using an independent test set, we extended our study to include a cohort of MDS patients collected at a separate institution. Because these subjects were studied using a closely related hematology analyzer, several parameters that were predictive in the first model were not available (RDW-SD, UWBC, MAF). To address this, we first retrained our classifier using only variables shared between the two instruments. Using our original training set for model construction, we determined that the new model yielded an AUC of 0.93 on the original test set. Given this comparable performance to the first model, we then applied the newly constructed model to the test set from a second institution. This second test set contained CBC data from 231 control subjects and 8 patients with MDS. The retrained classifier yielded a sensitivity of 75% and a specificity of 99.6% in this test set from the second institution, also generating an ROC AUC of 0.93 (Table 4).
| MDS random forest classifier | |||
|---|---|---|---|
| Diagnosis | Positive | Negative | |
| MDS | 6 | 2 | 75.0% Sensitivity |
| No MDS | 1 | 230 | 99.6% Specificity |
| 85.7% PPV | 99.1% NPV | ||
- MDS: myelodysplastic syndrome; PPV: positive predictive value; NPV: negative predictive value.
Discussion
Using a combination of standard CBC parameters, RUO parameters, and CPD data collected during routine automated CBC analysis, and patient age, we developed a random forest classifier that identifies patients that have MDS with relatively high sensitivity and specificity. By using an unselected patient population, as opposed to a group of patients with MDS and a control group, classifier performance was developed and evaluated in “real world” conditions with a heterogeneous mix of patients. Notably, standard clinical laboratory algorithms only flagged 25.0% of patients with MDS in the test set, whereas the classifier identified 85.0% of patients with MDS in the independent test set. Using the same methodology, we validated our findings in an independent dataset from a separate institution. A similar random forest classifier was able to identify patients with MDS in this dataset with an AUC of 0.93. Specificity was increased in this dataset versus the original (99.6% vs. 87.1%); similarly, the positive and negative predictive values are higher in the second dataset. The improved classifier performance in the second dataset is likely because the second dataset contained only patients with MDS and controls and has a higher prevalence of MDS, whereas the first dataset contains CBCs from a wide range of patients in a diverse outpatient setting.
The diagnostic sensitivity of the classifier at clinically feasible cutoff values is low for an ideal screening test; however, the classifier would not necessarily function as an independent screening test. Rather, it is designed to be utilized as an adjunct diagnostic that accompanies a commonly ordered laboratory test and does not require clinical suspicion of MDS. The cutoff values for the classifier are modifiable to find a balance between high sensitivity and increased peripheral smear reviews that is appropriate for a particular patient population. From a practical standpoint, data can be exported from the hematology analyzer after single CBCs or in batches, formatted (manually or in an automated fashion), and rapidly analyzed via the classifier.
In addition to serving as an adjunct diagnostic during routine CBC analysis, the classifier may also be useful in clinical settings with high suspicion for MDS. When bone marrow biopsies from study patients with clinical suspicion for MDS were examined, the classifier accurately categorized 5 of 6 patients in the independent test set of the original dataset. Use of the classifier in this specific clinical setting could serve as an objective assessment of the probability of MDS and assist in determining, which patients would benefit most from a bone marrow biopsy.
Identification of MDS through automated hematology analysis has been a subject of investigation for over 25 years 22. Several recent studies have demonstrated differences in automated CBC analysis parameters between patients with MDS and control patients with the hematology analyzer platform used in this study 20, 24 and other commercially available analyzers 25, 32, 33. Parameters with meaningful differences in patients with MDS have been relatively constant across studies within the same platform; Miguel et al. identified decreases in mean neutrophil light scatter and mean neutrophil conductivity in patients with MDS 24, whereas Haschke-Becher et al. identified an increased standard deviation of neutrophil conductivity as the best predictor of MDS (while also noting a less significant decrease in mean neutrophil conductivity) 20. In the current study, trends toward decreased mean neutrophil conductivity, decreased mean neutrophil light scatter, and increased mean volume scatter are noted; none of these trends achieves statistical significance after multiple testing correction, but fit the general pattern noted in prior studies with smaller, less heterogeneous study groups. These changes are also concordant with the expected morphologic differences seen in MDS, namely larger and hypogranular neutrophils.
One of the strengths of this study is the use of an unselected outpatient population during development and testing of the random forest classifier, rather than developing an algorithm to separate patients with known MDS from a selected “normal” control group. The use of an unselected population to develop a classifier has been previously performed, albeit with incomplete reporting of results and on a different hematology analyzer platform. Rocco et al. tested the performance of an algorithm used to identify patients with dysplastic features on a series of 80,000 samples from routine clinical practice, with 1.6% of patients identified by the algorithm as having MDS 26. Thirty-nine percentage of these patients were confirmed to have MDS clinically, whereas 61.0% of patients identified by the algorithm did not have MDS. Since clinical information was not reported on patients that were not flagged by this algorithm, the sensitivity and specificity of the algorithm cannot be calculated.
Although the proposed classifier performs well as a noninvasive adjunct screening method for MDS, it is important to note the relative contributions of the factors that are integrated into the model. The most statistically significant differences between the MDS group and remainder of the population are noted during univariate analysis of traditional CBC parameters such as hemoglobin, hematocrit, and RDW. The RUO parameters platelet distribution width (PDW) and standard deviation of the red cell distribution width (RDW-SD) demonstrate highest parameter discrimination within the classifier, followed by clinically reported CBC parameters. The RUO parameters of heightened red cell and platelet anisocytosis may reflect the coexistence of the MDS clone with normal cells. A similar classifier using these parameters may be applicable to other hematology analyzers.
CPD parameters are less discriminatory than RUO and clinically reported CBC parameters, but are useful to refine the classifier and provide added value. The effect of CPD parameters may be diluted in the larger dataset if there are patients with undiagnosed, early-stage MDS in the non-MDS population, since they would be expected to have CPD parameters that overlap with patients that have been identified clinically as having MDS. By utilizing ICD-9 codes for the clinical assessment of patients without MDS in the first dataset, some patients with MDS have likely been misclassified as non-MDS due to clerical errors in ICD-9 coding. Similarly, patients with chronic myelomonocytic leukemia, not included in the MDS ICD-9 group, may be present in the non-MDS population but have leukocytes with dysplastic CPD parameters. Finally, although extensive effort has been made to remove patients receiving chemotherapeutic agents from the study population, this may not have been completely successful. Although all of these scenarios are likely to be quite rare, the relatively small number of patients in the MDS group in the first dataset (n= 59) may magnify their effect.
In summary, we have developed a random forest classifier that identifies patients with MDS in two independent populations. Eighty-five percentage of patients with MDS were identified by the classifier in an unscreened outpatient population, as opposed to 25.0% with standard flagging algorithms. The classifier performed well (ROC AUC > 0.93) in both independent test sets. If validated with prospective studies and incorporated into routine clinical use, this classifier could aid early identification of patients with MDS.
Acknowledgments
The authors would like to thank Fernando P. Chaves, MD, Director of Global Scientific Affairs with Beckman Coulter, for scientific input and discussion on all aspects of this work, and Dr. Henk van Zaanen, Department of Internal Medicine, Sint Franciscus Gasthuis, Rotterdam, for help identifying the patients with MDS.




