Alteration of cohesin genes in myeloid diseases†
Conflict of interest: Nothing to report.
New genes involved in leukemogenesis, such as ASXL1 and TET2, have been identified recently using genomic analyses of DNA from patient samples. We have studied by array-comparative genomic hybridization (aCGH) a series of 167 samples including myelodysplastic syndromes, chronic myelomonocytic leukemias, and acute myeloid leukemias. We found a deletion of the RAD21 and STAG2 genes, which encode two components of the cohesin complex. We propose that these alterations may compromise the cohesin complex and its regulation of the transcription of genes.
Several new genes involved in myeloid leukemogenesis, including ASXL1, CBL, and TET2, have been identified following genomics studies using aCGH or single nucleotide polymorphism-arrays combined with sequence analyses [1-3]. Mutation analyses have identified other genes, such as IDH1 [4]. No alteration has been identified yet in many cases of malignant myeloid disease and many other genes remain to be discovered.
Using aCGH, we searched for genome alterations in a series of 167 malignant myeloid diseases. We describe here two cases with loss of genes encoding components of the same protein complex.
About two-thirds of the samples did not show any aCGH copy number aberrations (CNA): 41 of 63 MDSs (65%) (Supporting Information Table I), 37 of 53 CMMLs (∼70%) (Supporting Information Table II), 35 of the 51 AMLs (∼69%) (Supporting Information Table III). This high proportion of no-CNA profile in AMLs is probably due to our selection of cases with normal karyotype. In all cases, losses of whole or part of chromosome (e.g., 20q losses) were found in agreement with the results of the karyotyping (except for HD-0627 where the del20q could only be suspected on the aCGH profile).
| MDS | CMML | AML | Total | |
|---|---|---|---|---|
| Total number of studied samples | 63 | 53 | 51 | 167 |
| No CNA | 41 (65%) | 37 (69.8%) | 35 (68.6%) | 113 (67.7%) |
| ASXL1 loss | 1 | 0 | 1 | 2 |
| NF1 loss | 0 | 2 | 0 | 2 |
| RB1 loss | 0 | 2 | 0 | 2 |
| TET2 loss | 1 | 1 | 0 | 2 |
| Cohesin component loss | 0 | 1 | 1 | 2 |
Small deletions were more interesting. They were rare but indicative. We found a deletion of TET2 in one myelodysplastic syndrome (MDS) and in one chronic myelomonocytic leukemia (CMML), and a deletion of ASXL1 in one MDS and in one acute myeloid leukemia (AML; Table I). We also found deletions of NF1, RB1, RUNX1, and UTX. Some of these CNAs have been described in our previous reports [1, 5, 6]. We here focused on two particular CNAs.
CMML HD-0259 was diagnosed in 2007 in a 70 year-old patient with no particular antecedent. aCGH showed a small heterozygous deletion at 8q24 (chr8: 117768619-117969545 Mb), which comprised three genes including RAD21 (Fig. 1A). The CMML evolved in an M5 FAB AML (HD-0402) in 2008. aCGH again revealed the RAD21 loss but no additional alteration. The patient died in 2008, 6 months after acute transformation. AML HD-0120 (M6 FAB) was diagnosed in 2005 in a 55 year-old patient without any antecedent. aCGH detected a small deletion centered on the STAG2 gene, at Xq25 (chrX: 122787860-122970717 Mb) (Fig. 1B). The patient died in 2007, 5 months after relapse. In both cases, the karyotype did not show any abnormality and no other aCGH alteration was noticed.
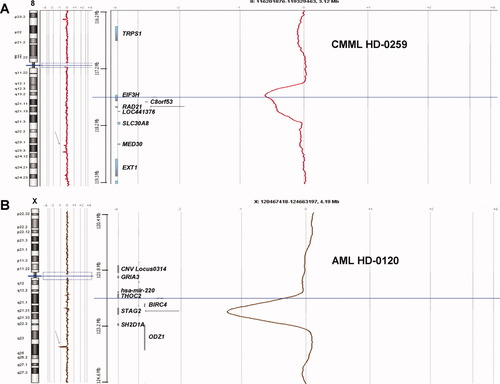
aCGH profiles showing loss of genes encoding cohesin subunits. A. CMML HD-0259 shows a loss of one copy of the RAD21 gene (arrows). B. AML HD-0120 shows the complete loss (male patient) of the STAG2 gene (arrows). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
HD-0120 had an IDH1 mutation (R132C). HD-0259 was the only CMML case with an NPM1 exon 12 mutation; a FLT3-ITD was additionally detected in the corresponding acute phase (HD-0402). There were no mutations in the other studied genes associated with leukemogenesis. We searched for mutations of RAD21 in 95 samples but did not find any.
At first look, deletions of RAD21 and STAG2 could appear as isolated cases with limited interest. However, close examination revealed the involvement of a new pathway of leukemogenesis. Indeed, the two genes happen to encode subunits of the same nuclear complex, cohesin [7]. Cohesin is made of four evolutionary-conserved core subunits, SMC1, SMC3, RAD21, and STAG1/STAG2 (Fig. 2), and plays a major role in chromosome biology [7]. Abnormal functioning of cohesin compromises faithful segregation of sister chromatids during cell division. Mutations in cohesin genes are responsible for the Cornelia de Lange syndrome (CdLs), a multisystem anomaly disorder with abnormal phenotypical and behavioral features, and may be associated with chromosome instability in colorectal cancer [7, 8]. However, we did not detect any aneusomy in the two cases. But cohesin has other roles. Cohesin participates in double-strand DNA break repair and long-distance regulation of gene expression. In zebrafish, monoallelic Rad21 inactivation downregulates Runx1 expression [9]. Cohesin is necessary for the function of the enhancer-blocking transcriptional insulator CTCF, which is an interactor of nucleophosmin and a positive regulator of tumor suppressor loci [10-12]. Cohesin can also acts on transcription independently of CTCF [13]. Thus, loss of RAD21 or STAG2 could participate in leukemogenesis through enhancement of inactivation of NPM1, RUNX1, and other tumor suppressor genes, such as RB1 or CDKN2A/B. Loss of RAD21 co-occurred with an NPM1 mutation in case HD-0259 showing that the two alterations can indeed cooperate. Abnormal repression of transcription could be a general mechanism of leukemogenesis. Alternatively, cohesin may silence oncogenes. The activity of cohesin/CTCF is regulated by polo kinase 1, which interacts with CEP170 [5] and is coordinated with epigenetic marks [10-12], which is in agreement with current views of oncogenesis [14, 15]. Thus, abnormal cohesin function may be involved in the same pathway as TET2 or ASXL1.
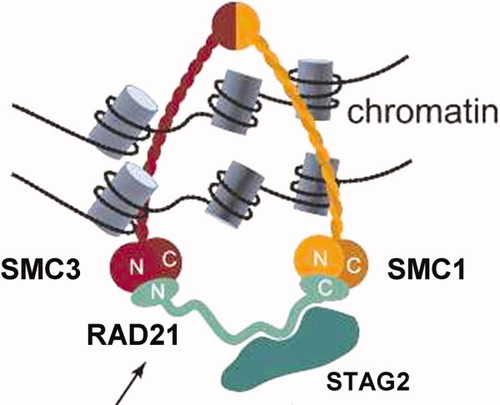
The mitotic cohesin complex is constituted of four core subunits (SMC1, SMC3, RAD21, and either STAG1 or STAG2), which embrace DNA and chromatin (Reproduced from Ref.7, with permission from Cold Spring Harbor Laboratory Press). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
To extend and validate our results, we surveyed published genomic studies of myeloid malignancies for alterations of cohesin genes. We found that one similar small deletion at 8q24 including RAD21 [16] and one at Xq25 including STAG2 [17] have been found in independent series of 157 and 86 AMLs, respectively. The method used to detect these CNAs allowed the conclusion that the deletions were acquired. In these studies, the involvement of cohesin was not suggested.
In array studies of myeloid diseases [1, 5, 6, 16, 17], recurrence is overall low but quite essential for identifying candidate target genes. Even if rare (1%), alterations of cohesin genes are truly recurrent and found in three independent series [16, 17 this work]. These results point for the first time to cohesin as a new player in leukemogenesis and contribute to establish the comprehensive catalog of pathogenic genes. They provide an incentive to look thoroughly for alterations (mutations?) in the genes encoding members of the protein complex and its associated network (e.g., CTCF), as well as to define the function of cohesin in hematopoiesis.
Materials and Methods
Panels of samples.
We collected samples from 63 MDS, 53 CMML, and 51 AML. According to the WHO classification, the MDS panel comprised five refractory anemia (RA), 13 RA with ring sideroblasts (RARS), six refractory cytopenia with multilineage dysplasia (RCMD), 16 RA with excess of blasts type 1 (RAEB1), 19 RA with excess of blasts type 2 (RAEB2), and 4 MDS-unclassified (MDS-U) cases. The 53 CMML panel comprised 24 myelodysplastic (MD) forms (leucocytosis count <13 × 109/L) and 29 myeloproliferative forms (leucocytosis count >13 × 109/L) (6). The AML panel comprised 39 cases of primary AMLs and 12 of secondary AMLs. It included 40 cases with normal karyotype and 10 cases with trisomy 8 and 1 with 20q deletion as a sole karyotypic abnormality. All patients signed an informed consent and the study was approved by our institutional review board.
Array-comparative genomic hybridization.
Genomic imbalances were analyzed by using 244K CGH Microarrays (Hu-244A, Agilent Technologies, Massy, France) as previously described [1, 5]. Some of the data have already been reported [1, 5, 6].
Sequencing.
We searched for mutations in 12 genes that have a demonstrated role in leukemogenesis: ASXL1, CBL, FLT3, IDH1, IDH2, JAK2, KRAS, NPM1, NRAS, RUNX1, TET2, and WT1. We also sequenced RAD21 in 47 MDS, 38 CMML, and 10 AML samples. Sequencing was done as described [1]. The sequence of all primers is available on simple request.
Author Contributions
JA did the genomic profiling and data analysis. JR and NC did the sequencing experiments and data analysis. VGB, AM, and MJM are biologists and were responsible for biological features collections. NV is physician in charge of the patients and clinical data. DB, MJM, and MC did project planning, integrated data analysis and manuscript writing.
References
Julien Rocquain*, Véronique Gelsi-Boyer* , José Adélaïde*, Anne Murati* , Nadine Carbuccia*, Norbert Vey §, Daniel Birnbaum*, Marie-Joelle Mozziconacci* , Max Chaffanet*, * Centre de Recherche en Cancérologie de Marseille, Laboratoire d'Oncologie Moléculaire, UMR891 Inserm, Institut Paoli-Calmettes, Marseille, France, Département de BioPathologie, Institut Paoli-Calmettes, Marseille, France, Faculté de Médecine, Université de la Méditerranée, Marseille, France, § Département d'Hématologie, Institut Paoli-Calmettes, Marseille, France.




