Genetic polymorphisms of the X-linked transcription factor forkhead box P3 predispose to synchronous secondary primary malignancy (SPM) of esophagus in head and neck squamous cell carcinoma patients
Funding information: National Cheng Kung University Hospital
Abstract
Patients with head and neck squamous cell carcinoma (HNSCC) are prone to develop second primary malignancy (SPM) at the esophagus synchronously, especially those with increased forkhead box P3 (FOXP3) positive regulatory T (Treg) cell infiltration in tumor tissue. We tested whether FOXP3 genetic polymorphisms can be associated with Treg cell infiltration and synchronous SPM in HNSCC patients. HNSCC patients received screening esophagogastroduodenoscopy during 2008 to 2016 were enrolled consecutively. After excluding and matching cigarette smoking and alcohol drinking status, the final cohort went on investigation. Three tag single nucleotide polymorphisms (tag SNPs) of the FOXP3 gene (rs2232365, rs3761548, and rs3761549) were selected from the HapMap database and genotyped by RT-PCR. FOXP3 immunohistochemistry was done to evaluate Treg cell amount in tumor tissues. A total of 270 male HNSCC patients were included, including 135 in the synchronous SPM (SynSPM group) and 135 in the HNSCC alone group. Compared with HNSCC alone group, synchronous SPM group had a HNSCC that was more commonly located at oro/hypopharynx (55.0% vs 39.6%, P = .012) and at earlier TNM stage with borderline significance (38.8% vs 29.0%, P = .124). In addition, carriage of G allele in rs3761549 of the FOXP3 gene significantly increased synchronous SPM risk (crude OR = 2.21, P = .009). After adjustment of HNSCC location and staging, rs3761549 G allele remained significantly associated with synchronous SPM risk (adjusted OR = 2.10, P = .024). Patients with G allele in rs3761549 also had higher FOXP3+ Treg cell infiltration in the tumor tissue than those with A allele (mean FOXP3+ cell number/mm2: 55.93 vs 36.39, P = .008). Male HNSCC patients with G allele in rs3761549 of the FOXP3 gene has a higher risk of synchronous SPM development and merit earlier screening endoscopy.
1 INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) is diagnosed in more than 600 thousand of patients globally, and resulted in more than 300 000 of cancer related deaths per year.1 The incidence rate was steadily high in recent years according to the cancer registration data worldwide.2, 3
Synchronous second primary malignancy (SPM) is defined as distinct, solid cancers diagnosed within 6 months of the diagnosis of index HNSCC.4 It is frequently detected in the upper aerodigestive tract, especially the esophagus, and leads to a poorer outcome in HNSCC patients.5-7 So, screening esophagogastroduodenoscopy (EGD) is recommended in HNSCC patients at high risk of developing esophageal synchronous SPM.8 Alcohol drinking and hypopharyngeal location of HNSCC were well known risk factors for synchronous SPM development in the esophagus.9 However, such environmental carcinogen exposure is common in HNSCC patients and we need other biomarkers to help select high risk candidates for screening EGD, which is time-consuming and suffering.10
FOXP3+CD25+CD4+ regulatory T (Treg) cell is essential and indispensable to suppress self-antigen-reacting T cells, a mechanism known as self-tolerance. However, tumor cells can also use this mechanism to escape from immune clearance.11 Tumor tissues from HNSCC patients with synchronous SPM had higher FOXP3+ Treg cell infiltration than those with HNSCC alone.12 This suggested Treg cell helps synchronous SPM development through immunosuppression. A key feature of Treg cell is to express FOXP3 protein responsible for its development and function.13
Single nucleotide polymorphism (SNP) is the genetic variation that occurs in more than 1% of the general population.14 It can change protein expression or function via alterations in transcription factor affinity, messenger RNA degradation, amino acid sequence, and messenger RNA splicing.15 SNP of FOXP3 has been reported to be an important risk factor for several kinds of solid tumors, including HNSCC.13 This study aimed to investigate whether FOXP3 SNP is associated with an increased Treg cell infiltration in the tumor tissue and development of synchronous SPM in HNSCC patients.
2 MATERIALS AND METHODS
2.1 Patient enrollment
We consecutive enrolled HNSCC patients who received screening EGD for synchronous cancer in the esophagus in National Cheng Kung University Hospital from 2009 to 2016 and E-Da Hospital from 2008 to 2011. Patient whose esophageal squamous cell carcinoma noted within 6 months of diagnosis of HNSCC was classified as synchronous SPM group (case). The others were classified as HNSCC group (control). Total 476 patients were enrolled during this period, among them 180 patients belonged to synchronous SPM group and the other 296 patients were HNSCC group. Patients' baseline characteristics, disease status such as head and neck cancer location, stage (according to TNM staging system), survival time and environmental carcinogen exposure (cigarette smoking, alcohol drinking and betel nuts chewing) were all recorded. The institutes' review board and ethics committees of both E-Da Hospital (EMRP35102N) and National Cheng Kung University Hospital (A-ER-104-350) had approved the study.
After enrollment, we excluded patients who had chemoradiotherapy or any kind of tumor treatment prior to screen EGD to avoid its interference on esophageal synchronous SPM development (n = 132). Due to small case numbers of female patient (n = 3) and the difference in genotyping in a gene located on X chromosome of female, we excluded all three female patients. Patients without available DNA were also excluded (n = 22). To control the known environmental risk factors of synchronous SPM, such as cigarette smoking and alcohol drinking, these two factors were matched in the case and control groups. Betel nut chewing state was not matched because there is no evidence of its role in esophageal synchronous SPM development currently. As a result, a total of 270 patients, 135 in each group were included for final analyses.
2.2 Tag single nucleotide polymorphism selection
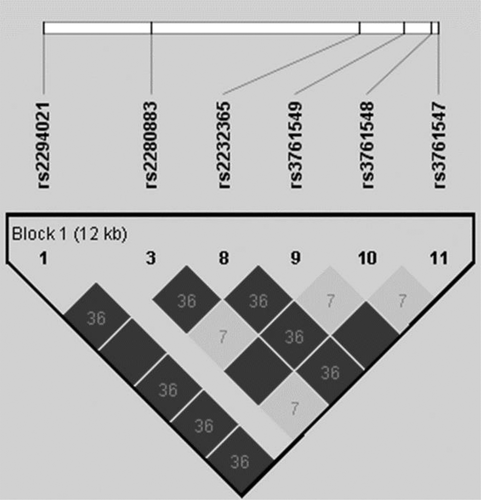
The International HapMap database release #27 (http://hapmap.ncbi.nlm.nih.gov/index.html.en) of Chinese Han Beijing (CHB) group was used to select tag SNPs in the FOXP3 gene region from chromosome X at nucleotide position 49 250 436 to 49 265 738, with a minor allele frequency > 5%. Under this condition, total seven tag SNPs (rs2294021, rs2280883, rs2232365, rs3761547, rs3761548, and rs3761549) were Listed. After confirming the LD (linkage disequilibrium), 3 tag SNPs (rs2232365, rs3761548, and rs3761549) were enough to represent all genotypes of FOXP3 gene. The detail LD plot of CHB group in the HapMap database is shown in Figure 1.
2.3 DNA extraction and genotyping
All the enrolled patients' blood was collected after screening esophagogastroduodenoscopy and obtaining inform consent. Genomic DNA was extracted from lymphocyte using a DNA isolation kit (QIAGEN, Germany) with standard protocol provided by the user's operation manual. The SNPs were detected by primer extension analysis using endpoint TaqMan assays (Applied Biosystems, Warrington, UK) in 96-well arrays, and genotypes were subsequently read on a StepOne Real Time PCR Detector (Applied Biosystems). Reactions were carried out using standard conditions offered by the manufacturer.
2.4 FOXP3+ Treg cell density in the tumor tissue by Immunohistochemistry
Paraffin-imbedded tumor tissues were sectioned in 0.4 μm thickness for histological examination and immunohistochemistry. During immunohistochemistry, the samples were deparaffinized by xylene and rehydrated with 95% and 75% alcohol subsequently. Then the tissue protein was reactivated by autoclaving with DakoCytomation Target Retrieval Solution for 8 minutes. The endogenous peroxidase activity was quenched by immersing the specimens in 0.3% hydrogen peroxide for 10 minutes. Nonspecific binding sites were saturated with SuperBlock Blocking Buffer (Thermo Scientific, IL). Tissue sections were treated with primary antibody against FOXP3 (clone 246A/E7; Abcam, Cambridge, UK) at a dilution of 1:40. A pathologist blinded to patients' clinical information quantified the number of FOXP3+ Treg cell by analyzing five different fields (×200). Each observed field represent an area of 0.785 mm2. The density (cells/mm2) of intra-tumor infiltrating Treg cell was defined as the mean FOXP3+ lymphocyte numbers in five fields divided by the observed area.
2.5 Statistical analyses
SPSS software (version 20.0, IBM Corporation, Armonk, NY) was used for statistical analyses of the data. The differences of baseline characteristics between the two groups were examined with Student t test or χ2 test as appropriate. For SNPs analysis, we conducted logistic regression to calculate both the crude odds ratio and adjusted odds ratio of each SNP. Adjusted odds ratio (AORs) was obtained to find independent risky SNPs after correcting factors that had significant or borderline significant difference (P < .2). The difference of FOXP3+ Treg cell density in the tumor tissue between groups was analyzed with Student t test.
3 RESULT
3.1 Baseline characteristics of the enrolled patients
A total of 270 male HNSCC patients with matched alcohol and cigarette exposure were included for final analyses, including 135 with synchronous SPM (synchronous SPM group) and 135 without (HNSCC alone group). Their baseline characteristics were listed in Table 1. Synchronous SPM group more commonly had their HNSCC occurred at oropharynx or hypopharynx compared to HNSCC alone group (55.0% vs 39.6%, P = .012). Synchronous SPM group also had a lower HNSCC stage, although not statistically significant. Age and betel nut chewing were not significantly different between the two groups. Factors that had a P value less than 0.2 were adjusted while analyzing the association between FOXP3 genetic polymorphism and synchronous SPM risk.
| HNSCC (n = 135) | Syn SPM (n = 135) | P value | |
|---|---|---|---|
Age years, mean ± SD (range) |
53.58 ± 10.01 (22-77) | 52.93 ± 8.29 (37-76) | .561 |
| HNSCC location | |||
| Other sites (%) | 81 (60.4) | 58 (45.0) | .012 |
| Oro/hypopharynx (%) | 53 (39.6) | 71 (55.0) | |
| HNSCC stage | |||
| 0-II (%) | 32 (29.0) | 45 (38.8) | .124 |
| III-IV (%) | 78 (71.0) | 71 (61.2) | |
| Betel nut chewing | |||
| No (%) | 117 (86.7) | 113 (85.0) | .689 |
| Yes (%) | 18 (13.3) | 20 (15.0) | |
| Cigarette smokinga | |||
| No (%) | 3 (2.2) | 3 (2.2) | 1.000 |
| Yes (%) | 132 (97.8) | 132 (97.8) | |
| Alcohol drinkinga | |||
| No (%) | 5 (3.7) | 4 (3.0) | .735 |
| Yes (%) | 130 (96.3) | 131 (97.0) | |
- Abbreviations: HNSCC, head and neck squamous cell carcinoma; Syn SPM, Synchronous second primary malignancy.
- [a] Smoking and alcohol drinking state were already matched before analysis.
3.2 The association between FOXP3 genotypes and synchronous SPM risk
Table 2 compared the genotype frequencies of the three FOXP3 tag SNPs (rs2232365, rs3761548, and rs3761549) in patients with and without synchronous SPM. This study used major allele of the study population as the referent allele for the three tag SNPs. The major allele in our study population was T for rs2232365, G for rs3761548, and G for rs3761549, the same as HapMap database. Because FOXP3 gene is located on the chromosome X and all the enrolled patients were male, each tag SNP only had a single allele during genotyping. As shown in Table 2, patients with esophageal synchronous SPM had a significantly higher frequency of G genotype in rs3761549 (84.4% vs 71.1%, P = .009) and a trend of increased frequency of T genotype in rs3761548 (25.2% vs 17%, P = .101), compared to patients with HNSCC alone. Patients with G genotype in rs3761549 had a 2.21-fold risk (95% CI, 1.22-4.00, P = .009) of developing esophageal synchronous SPM compared to those with A genotype. After adjusting HNSCC location and stage, carriage of G genotype in rs3761549 still had a 2.10-fold risk (95% CI, 1.11-4.00, P = .024) of developing esophageal synchronous SPM. These data suggest G genotype in rs3761549 is an independent risk factor for developing esophageal synchronous SPM in HNSCC patients. Other SNPs failed to reach statistical significance.
| HNSCC, n (%) | Syn SPM, n (%) | Crude OR (95% CI) | P value | Adjust ORa (95% CI) | P value | |
|---|---|---|---|---|---|---|
| rs2232365 | ||||||
| T allele | 72 (53.3) | 80 (59.3) | 1 | .326 | 1 | .320 |
| C allele | 63 (46.7) | 55 (40.7) | 0.79 (0.49-1.27) | 0.76 (0.45-1.30) | ||
| rs3761548 | ||||||
| G allele | 112 (83.0) | 101 (74.8) | 1 | .101 | 1 | .179 |
| T allele | 23 (17.0) | 34 (25.2) | 1.64 (0.91-2.97) | 1.55 (0.82-2.94) | ||
| rs3761549 | ||||||
| G allele | 96 (71.1) | 114 (84.4) | 1 | .009 | 1 | .024 |
| A allele | 39 (28.9) | 21 (15.6) | 0.46 (0.25-0.82) | 0.48 (0.25-0.91) | ||
| G vs A | 2.21 (1.22-4.00) | .009 | 2.10 (1.11-4.00) | .024 | ||
- Abbreviations: HNSCC: head and neck squamous cell carcinoma, Syn SPM: Synchronous second primary malignancy.
- [a] Adjust HNSCC tumor location and stage.
3.3 Abundance of FOXP3+ regulatory T cell infiltration in the tumor tissues from patients carrying risky genotype
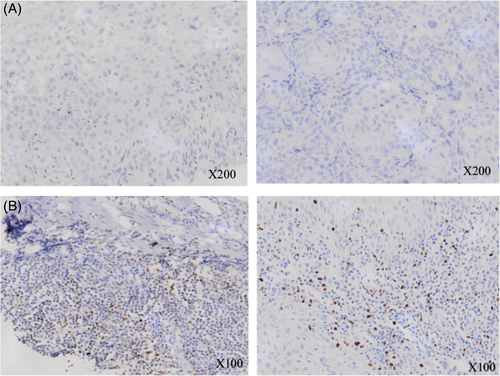
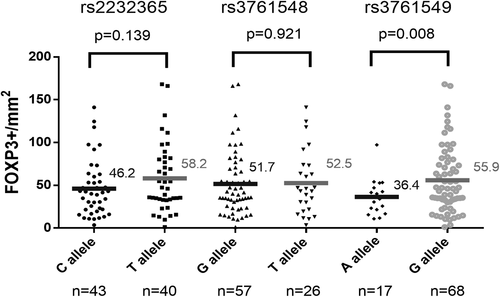
We further determined whether genetic polymorphism in FOXP3 can affect FOXP3+ Treg cell infiltration in tumor tissue. There were 83 tumor tissues available for immunohistochemical (IHC) staining (35 from esophageal synchronous SPM and the other 48 from HNSCC). Figure 2 shows the representative pictures of FOXP3 IHC stain in the studied patients. As shown in Figure 3, tumor tissues from patients carrying G allele in rs3761549 had significantly higher amount of FOXP3+ Treg cell infiltration compared to those from patients carrying A allele (55.93 vs 36.40 cells/mm2, P = .008). The FOXP3+ Treg cell density showed no significant difference between tumors from patients with different genotypes in the other two tag SNPs.
4 DISCUSSION
In this hospital based, case-control study, after matching cigarette smoking and alcohol drinking state, we found oro/hypopharyngeal location of HNSCC was significantly associated with increased esophageal synchronous SPM risk. In addition, G allele in rs3761549 of FOXP3 gene also independently increased esophageal synchronous SPM risk. We further strengthened our findings by counting FOXP3+ Treg cell number in the tumor tissue as a functional assessment of FOXP3 genetic polymorphism. And significant increase in FOXP3+ Treg cell number was found in tumor tissues from patients carrying G allele in rs3761549.
We found rs3761549 G allele of FOXP3 gene is the risk genotype for synchronous SPM development. Previous studies also reported that FOXP3 genetic polymorphism affected the risk of several types of cancers in different ethics. A study in Chinese-Han Beijing (CHB) population found rs3761549 G allele was also a risky allele for hepatoma.16 Another study found rs3761548 T allele increased the risk of lung cancer, but rs3761549 was not genotyped in that study.17 Similarly, we found rs3761548 T allele was marginally associated with increased synchronous SPM risk (crude OR = 1.64, P = .101). Further study with a bigger case number is needed to clarify the role of this locus.
Our study group had previously found that FOXP3+ Treg cell infiltration was increased in synchronous SPM tumor tissue compared to the tumor tissues from patients with HNSCC or esophageal SCC alone.12 The study showed that infiltration of Treg > 34.6 cells/mm2 resulted in a 6.13-fold risk for developing synchronous SPM, which was higher than the risk of alcohol drinking and cigarette smoking. The study results suggested that cellular immunosuppression may lead to synchronous SPM development. In this study, we further showed that G allele in in rs3761549 of the FOXP3 gene correlated with higher Treg cell infiltration in the tumor tissue. Thus, the FOXP3 genetic polymorphism may predispose to synchronous SPM development via stimulation of Treg cell to inhibit anti-tumor immunity.
FOXP3+ Treg cell plays an important role in self-tolerance and its linkage to human immune system disease is well established.18, 19 In human malignant disease, emerging evidence suggest that FOXP3+ Treg cell is also important for immune escape of cancers.20, 21 Cancer cell may escape from immune cell attack if there is abundant infiltration of Treg cell in the tumor tissue. Therefore, it is reasonable that FOXP3 genetic polymorphism may increase Treg cell maturation via increased FOXP3 protein production, and aid in synchronous SPM development. In NCBI database, rs3761549 locates in the intronic region of FOXP3 gene. Theoretically, intron does not involve in protein coding and may not alter FOXP3 protein structure. However, emerging evidence revealed that intron may affect gene splicing, transcription factor binding, and messenger RNA degradation.15 In this study, we provided evidence that the risky genotype (G allele in rs3761549) was associated with increased Treg cell density. Further functional studies are needed to prove that G allele in rs3761549 can really increase FOXP3 protein expression.
This study still had some limitations. First, although we enrolled every patient who received screening EGD for esophageal synchronous SPM, our cohort had a higher frequency of esophageal synchronous SPM than that observed in previous reports. It is possible that otolaryngologist referred patients with high suspicious of synchronous esophageal cancer for EGD screening. However, we matched the risk factors and performed adjusted analyses which can eliminate the influence of confounding factors to reinforce the role of genetic polymorphisms. Second, the selected SNPs in this study all locate in the intronic region. The causal relationship between gene and protein expression/function in molecular level remained to be confirmed. However, the immunohistochemistry stain in our study proved the possible relationship indirectly.
In conclusion, G allele in rs3761549 has an effect in synchronous SPM development in male HNSCC patients and correlate to have a higher FOXP3+ Treg infiltration in tumor tissue. Male HNSCC patients who carry G allele in rs3761549 merit early screening and aggressive EGD surveillance to find esophageal synchronous SPM.