Cyclic Stiffness Modulation of Cell-Laden Protein–Polymer Hydrogels in Response to User-Specified Stimuli Including Light
Abstract
Although mechanical signals presented by the extracellular matrix are known to regulate many essential cell functions, the specific effects of these interactions, particularly in response to dynamic and heterogeneous cues, remain largely unknown. Here, a modular semisynthetic approach is introduced to create protein–polymer hydrogel biomaterials that undergo reversible stiffening in response to user-specified inputs. Employing a novel dual-chemoenzymatic modification strategy, fusion protein-based gel crosslinkers are created that exhibit stimuli-dependent intramolecular association. Linkers based on calmodulin yield calcium-sensitive materials, while those containing the photosensitive light, oxygen, and voltage sensing domain 2 (LOV2) protein give phototunable constructs whose moduli can be cycled on demand with spatiotemporal control about living cells. These unique materials are exploited to demonstrate the significant role that cyclic mechanical loading plays on fibroblast-to-myofibroblast transdifferentiation in 3D space. The moduli-switchable materials should prove useful for studies in mechanobiology, providing new avenues to probe and direct matrix-driven changes in 4D cell physiology.
There is a growing appreciation for the large role that mechanical signals presented by the local extracellular matrix (ECM) have on cell function. Through direct interaction with physical cues presented in the cellular ECM, these external mechanical signals are translated into internal biochemical responses that govern gene expression and cell fate decisions.1 Seminal findings in the field of cellular mechanotransduction have demonstrated that matrix stiffness alone can drive changes in essential processes including attachment, cytoskeletal organization, migration, proliferation, and differentiation.2-4 More recently, it has been observed that cells possess mechanical “memory,” storing information about past physical culture conditions to influence future behaviors.5 These findings represent landmark observations that are rapidly changing standard practices in molecular biology and stem cell culture.
Efforts to elucidate the specific effects that ECM stiffness and elasticity have on cell physiology have been performed almost exclusively using static biomaterials. While these studies have provided invaluable insight into the critical roles in which ECM stiffness regulates cellular fate, such simple systems fail to emulate biophysical dynamics known to accompany tissue/organ development, regeneration, and disease progression. Strategies to probe the biological effects of evolving tissue compliance have yielded a variety of synthetic cell culture platforms that can either soften or stiffen over time.6 Though constructs that undergo spontaneous or cell-mediated transitions have proven beneficial in several applications, those that can be modified on demand are critical for probing biophysical responses at well-defined times.7 Light-mediated material alteration has proven particularly beneficial in this regard as it uniquely grants near-instantaneous and spatiotemporal control over matrix stiffness in a potentially biocompatible manner.8 Material secondary photo-crosslinking enables one-way stiffening,9-14 while photodegradation provides for irreversible softening,15-19 in the presence of live cells.
Beyond unidirectional elasticity changes, cells also experience cyclic loading that periodically and reversibly alter local ECM rigidity; pulsatile flow associated with the circulatory system places cyclic loads on cells virtually everywhere throughout the body (timescale of seconds), just as tissues exhibit sequential stiffening and softening during wound healing (days to weeks). Despite significant interest in understanding these fundamental biological processes,6, 20-23 materials capable of recapitulating such dynamic and reversible stiffening remain largely undeveloped. Though several reversibly compliant biomaterials sensitive to a variety of stimuli have been reported,24-36 none have proven capable of examining changes in 3D cell response to cyclic moduli alteration, for example, owing to cytotoxic conditions required for material formulation/modification or by unavoidable interference in material modulation chemistries by nonspecific interactions with cell culture media components. Moreover, though local heterogeneities guide cell fate anisotropically within tissues, spatial control over reversible stiffening in materials compatible with 3D cell culture has not been previously demonstrated.
Here, we introduce a generalizable strategy to create hydrogel biomaterials whose moduli can be reversibly switched in response to user-specified inputs, potentially with spatial control, and then use these unique materials to probe the effects of cyclic mechanical loading on encapsulated cell fate. Wedding changes of biomolecular conformation to those in gel crosslinking density, we genetically fuse stimuli-responsive proteins with their stimuli-dependent binding partners (Figure 1a). In the presence of the external stimulus, the responsive protein undergoes a conformational change that promotes binding to its fused partner and a shortened end-to-end length. Following removal of this stimulus (or introduction of another), intramolecular protein–protein interactions are destroyed and original protein conformation is restored. When these fusion proteins are incorporated into the hydrogel crosslinker itself, molecular shortening translates to an elastic stiffening of the network, while the extended conformation yields a softer material (Figure 1b). As stimuli introduction and removal can be repeated ad infinitum, fully reversible control over biomaterial mechanical properties is readily obtained. Since several hundred proteins have been determined to undergo stimuli-dependent binding with known partners,37 many smart moduli-switchable materials responsive to distinct classes of stimuli (e.g., ligands, pH, temperature, ions, light) can be readily produced.
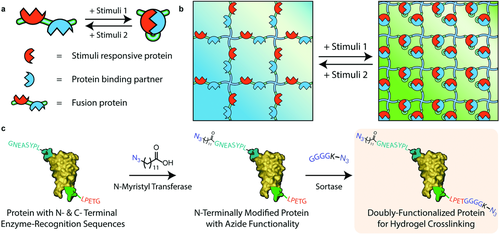
To generate stiffness-cyclable hydrogels, we perform a step-growth polymerization via a strain-promoted azide–alkyne cycloaddition (SPAAC) between a four-arm poly(ethylene glycol) (PEG) tetrabicyclononyne (tetraBCN) (Mn ≈ 20 kDa), a linear PEG diazide (N3PEGN3, Mn ≈ 3.5 kDa), and a fusion protein uniquely end-functionalized with reactive azides (N3) at both the N- and C-termini (Methods S1–S4, Supporting Information). Though SPAAC's bioorthogonality has been previously utilized to formulate cell-laden peptide–polymer hydrogels that exhibit idealized structure and well-defined composition,18, 38-42 this chemistry has not yet been exploited to create protein–polymer networks. Furthermore, precisely installed bioorthogonal handles enable almost any protein species to be used as a crosslinker without the loss-of-activity typical of site-directed mutagenesis or the uncontrollable heterogeneity that accompanies statistical functionalization of endogenous amino acid residues.43 By varying the ratio of the azide-modified crosslinkers (i.e., N3PEGN3 vs fusion protein) present during gelation, the maximal extent of material response can be precisely controlled.
End-modified protein crosslinkers are prepared through a novel orthogonal pair of chemoenzymatic modifications (Figure 1c). N-terminal labeling is achieved by N-myristoyl transferase (NMT), which promotes cotranslational fatty acylation on proteins bearing the “GXXXS/T” signature sequence (where X is any amino acid). NMT tolerates many synthetic analogs of its natural myristic acid substrate; enzymatic modification with 12-azidododecanoic acid (12-ADA, Method S5, Supporting Information) yields site-specific installation of azide functionality at the N-terminus.44-46 To append bio-orthogonal handles onto the C-terminus of proteins, we exploit a versatile sortase-mediated transpeptidation reaction.47, 48 Staphylococcus aureus sortase A is a calcium-assisted transpeptidase that catalyzes the cleavage of a C-terminal sorting signal “LPXTG” with the concomitant amide linkage of a polyglycine probe and the target protein. “Sortagging” with an azide-bearing oligoglycine permits efficient C-terminal installation of azido functionality (Method S6, Supporting Information). Though both NMT and sortase have each been used in isolation to introduce a single reactive azide onto either the N- or C-terminus of a protein,44-46, 49 their combined usage has not been demonstrated in literature. In fact, modification of a single protein by multiple chemoenzymatic reactions has not yet been established. Here, we demonstrate that recombinant proteins bearing both enzymatic recognition sequences can be readily prepared through standard molecular cloning and expression techniques. As chemoenzymatic transformations proceed with exceptionally high labeling efficiencies,50 we hypothesized that uniform populations of dually functionalized protein-based material crosslinkers could be created from virtually any monomeric or fused species, and that they could function uniquely as macromolecular precursors for the controlled step-growth polymerization of protein–polymer hydrogels.
Having identified a strategy to create stiffness-cyclable hydrogels using dually modified responsive proteins, we focused our initial efforts on calmodulin (CaM), a calcium-sensitive protein whose stimuli sensitivity has been previously utilized to make dynamic materials.24, 25 Taking lead from an expansive body of genetically encoded calcium sensors,51-54 we created a fusion of CaM and the M13 peptide fragment of myosin light chain kinase that could be dually modified for gel crosslinking (Method S7, Supporting Information). When Ca2+ is present, CaM undergoes a conformation change that permits high-affinity binding with M13; when the ion is chelated away, CaM structure and intramolecular association relaxes (Figure 2a). Following expression and modification by NMT and sortase, the end-modified N3CaMM13N3 complex was obtained in good yield (≈5 mg L−1 culture, nonoptimized expression, Method S8, Supporting Information). Whole-protein mass spectrometry revealed exceptionally high sample purity and quantitative functionalization at both termini (Figure 2b, Method S9 and Tables S1 and S2 (Supporting Information)); the observed mass for N3CaMM13N3 (22 807 Da) correlated well with its expected value (22 805 Da). Far-UV circular dichroism confirmed responsiveness of the complex to calcium (Figure 2c), consistent with similar analysis performed on purified CaM,55 where α-helical content increases upon Ca2+ binding, as indicated by decreasing ellipticity at 222 nm.56 To verify that the introduced azides were functionally accessible and maintained reactivity by SPAAC, we utilized a sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel shift assay57 involving C-/N-terminal PEGylation by BCN-monofunctionalized methoxy-PEG (mPEGBCN, Mn ≈ 10 kDa, Method S10, Supporting Information) (Figure 2d); the near-complete disappearance of the starting protein band, accompanied by the simultaneous appearance of a new band upshifted by the average molecular weight of the correct number of PEG chains (0, 1, or 2, depending on condition), indicates the ability to install reactive azides site specifically onto CaMM13 proteins using NMT and/or sortase. Collectively, these results demonstrate the successful generation of a bioresponsive fusion protein-based crosslinker, as well as the first utilization of two different chemoenzymatic reactions to modify a single recombinant protein.
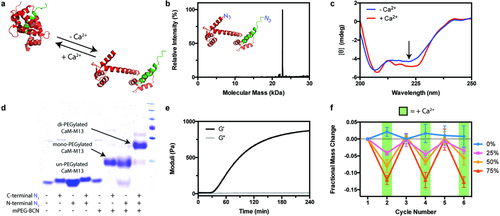
Calcium-responsive hydrogel networks were formed based on the SPAAC reaction of PEGtetraBCN (2 × 10−3 m), N3PEGN3 (2 × 10−3 m), and N3CaMM13N3 (2 × 10−3 m). Dynamic time-sweep rheological experiments indicated gelation ≈30 min after mixing, estimated by the crossover point between the elastic (G′) and storage moduli (G″), and a final G′ of 920 ± 50 Pa and G″ of 15 ± 5 Pa at t ≈ 5 h (Figure 2e). To assess changes in network composition in response to free-ion treatment, we examined cycled changes of mass swelling ratios in the presence or absence of calcium. When incubated with Ca2+, materials crosslinked with 50 mol% CaMM13 exhibit a 6.8 ± 1.5% decrease in swollen gel mass, reflecting the overall network constriction and a net loss in equilibrium water retention that accompanies CaM association with M13 (Figure 2f). Varying the ratio of N3PEGN3 to N3CaMM13N3 while keeping their total concentration constant (4 × 10−3 m) gave materials with scaled responsiveness; gels formed with 0, 25, 50, and 75 mol% CaMM13, respectively, exhibited 1.5 ± 2.3%, 4.1 ± 0.9%, 6.8 ± 1.5%, and 12.4 ± 1.7% Ca2+-promoted decrease in mass swelling. Based on Flory–Rehner polymer network theory,58 which predicts that swelling ratio (Q) of highly swollen materials scales with crosslinking density (ρx) as Q ∼ρx−3/5 and shear moduli (G) as G ∼ρx6/5, Ca2+-promoted decrease in swelling observed for the 25, 50, and 75 mol% CaMM13, respectively, results in a 8.7 ± 2.0%, 15 ± 3.8%, and 30 ± 4.8% relative gel stiffening. In all cases, initial hydrogel mass was restored following egtazic acid (EGTA)-mediated chelation of free Ca2+. Alternate treatments of Ca2+ and EGTA gave rise to reproducible material cyclability. We expect that the extent of hydrogel stiffening could be further tuned by varying the distance change that accompanies the fusion protein's conformational switch, which could be accomplished by varying molecular weight of multiarm PEG or through addition of spacers between protein binding partners.
Though the CaMM13 system proved useful in validating our overall fusion protein-based approach, its utility in probing cell fate response to cyclic stiffening is limited: 1) alterations to network mechanics rely on slow molecular diffusion events, preventing rapid control over material switchability; 2) calcium signaling is involved in almost all aspects of cellular life,59 rendering it impossible to alter network parameters without also perturbing biological function; and 3) spatial control over network mechanics is not readily possible. To address each of these limitations, we identified the photoswitchable LOV2Jα binding pair for use as a stimuli-responsive fusion protein. Recently popularized by the optogenetics community,60-63 the light, oxygen, and voltage sensing domain 2 (LOV2) is a plant-derived photoreceptor protein with a high binding affinity toward the C-terminal Jα helix.64 In the presence of blue light (λ = 470 nm), LOV2 initiates a photochemical reaction with a flavin mononucleotide chromophore (FMN), forming a covalent linkage between FMN and a critical cysteine residue on LOV2, that results in a displacement of the Jα domain and a large conformational shift (end-to-end changes estimated to be on the order of tens of Angstroms). This change occurs rapidly in response to low levels of blue light, and reverses quickly in the dark (Figure 3a). As cells are considered fully tolerant to light with wavelengths ≥ 365 nm,65, 66 there is no concern over UV-induced DNA damage when visible light sources are employed. We anticipated that utilization of LOV2Jα as a gel crosslinker would enable unique photoreversible and spatiotemporal control over hydrogel stiffness in the presence of live cells.
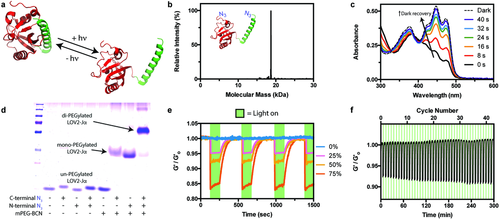
Expression of N3LOV2JαN3 yielded a pure species with quantitative functionalization, as confirmed by whole-protein mass spectrometry (Figure 3b, Methods S7–S9 and Tables S1 and S2 (Supporting Information)); the observed mass for N3LOV2JαN3 (18 841 Da) correlated well with its expected value (18 848 Da). The diazide protein exhibited the near-instantaneous shift in absorbance in response to blue light (λ = 470 nm), followed by a rapid dark recovery (t1/2 = 12 ± 5 s, high standard deviation based on equipment sampling frequency limitations) characteristic to the unmodified LOV2,67 indicating that the protein is properly folded and that the FMN chromophore survives purification (Figure 3c and Figure S1 (Supporting Information)). Gel shift assays performed with mPEGBCN confirmed that azides affixed at both the N- and C-termini of LOV2Jα were accessible and maintained reactivity, as well as successful generation of a light-responsive protein-based crosslinker (Figure 3d).
Photoswitchable gels were formed by reaction of PEGtetraBCN (2 × 10−3 m), N3PEGN3 (2 × 10−3 m), and N3LOV2JαN3 (2 × 10−3 m). In situ rheometry indicated that hydrogels formed on similar timescales and with comparable mechanics as CaMM13-based materials. Photorheometry studies revealed that fully formed gels exhibited rapid softening (t1/2 = 0.9 ± 0.2 s) in response to mild visible light (λ = 470 nm, 10 mW cm−2) (Figure 3e). Varying the ratio of N3PEGN3 to N3LOV2JαN3 while keeping their total concentration constant (4 × 10−3 m) gave materials with scaled responsiveness; gels formed with 0, 25, 50, and 75 mol% LOV2Jα, respectively, exhibited 0.0 ± 0.1%, 4 ± 1%, 8 ± 1%, 15 ± 1% photomediated moduli deflation. The softened state was retained until light exposure was ceased, upon which full material recovery was observed (t1/2 = 35 ± 1 s). As expected, the timescale for gel recovery is similar but slightly slower (approximately threefold) than that of LOV2's covalent separation from FMN, as presumably some time is required for LOV2Jα binding partners to reassociate following LOV2's conformational change. Characteristic timescales for material softening and stiffening were statistically indistinguishable for each LOV2Jα gel composition. We note that LOV2's temporary “bleaching” following exposure at wavelengths used to trigger its conformational change (Figure 3c) minimizes light attenuation and enables rapid modulation of relatively thick materials. In comparison with reported gels that soften through photoinduced intermolecular dissociation, particularly those employing high concentrations of photoactive moieties with large molar absorptivity,28, 34-36 LOV2Jα gels can be uniformly softened throughout the bulk material without inducing surface erosion. By shuttering light exposure to gels, full cyclability was obtained for all materials. Negligible material fatigue was observed for 50 mol% LOV2Jα gels cycled >40 times over 5 h (Figure 3f and Figure S2 (Supporting Information)). Such moduli cyclability, made possible by rapid molecular rearrangement events, has not been demonstrated previously with any other biocompatible system.
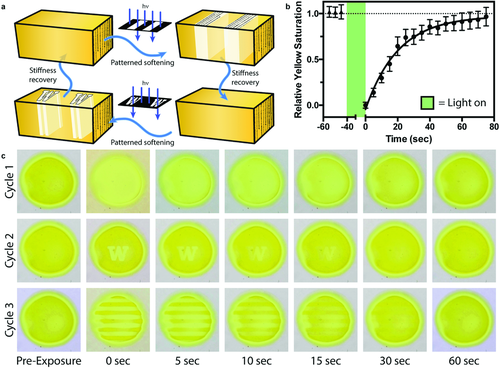
In addition to providing unprecedented temporal and cycled control over matrix properties, directed light exposure can be used to confine LOV2Jα gel softening to user-defined physical locations (Figure 4a). As the fast gel transition times and light responsiveness prevent localized mechanical characterization by many conventional techniques (e.g., atomic force microscopy, nanoindentation, microrheology), we took advantage of the distinct visible color shift that accompanies LOV2 photoactivation to monitor local stiffness changes within gels (Figure 3c); stiffer materials where LOV2 is noncovalently bound to FMN but is associated with Jα appear yellow under ambient light, while softer substrates in which LOV2 is covalently bound to FMN and dissociated from Jα are visually clearer. Image analysis of gel color following blue light flood exposure (λ = 470 nm, 1 mW cm−2, 30 s) revealed a dark recovery rate (t1/2 = 14 ± 3 s, Figure 4b) consistent with in-solution LOV2Jα analysis. Patterned regions of gel softening were created by traditional photolithographic techniques, whereby collimated light was shone through a photomask onto the surface of an optically thin sample. Gels were imaged before and after photopatterning; temporary changes in gel color and moduli matched masked-defined shapes (Figure 4c and Movies S1–S3 (Supporting Information)). Upon complete stiffness recovery, sequential patterning was performed using the same gel and a different photomask. Results highlight the platform's unique spatiotemporal control over material compliance as well as its reversibility, where patterning can be repeated many times over to soften different material regions as desired. Furthermore, laser-scanning lithographic technique enables gels to be reversibly softened with micrometer-scale features that approach the size of single cells (Figures S3 and S4, Supporting Information).
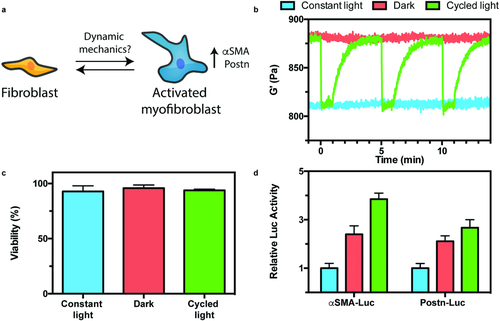
Using our reversibly compliant LOV2Jα hydrogels, we sought to establish the first understanding concerning the role of cyclic mechanical loading on myofibroblast activation in 3D. Upon injury, fibroblasts transdifferentiate into myofibroblasts that promote ECM remodeling.68 This activation can be regulated chemically through cytokine stimulation as well as through physical interactions with matrix substrates, and is associated with increased smooth muscle α-actin (αSMA) and periostin (Postn) gene expression.69 Though material stiffness and cyclic stretching of flexible 2D materials (which have not yet been shown to support cell encapsulation) can drive changes in enhanced activation,70, 71 material limitations have prevented investigation of the effects of dynamic mechanical loading on 3D myofibroblast transformation (Figure 5a). To probe these effects, NIH/3T3 fibroblasts were encapsulated (5 × 106 cells mL−1) in 50 mol% LOV2Jα hydrogels (a composition initially chosen to minimize protein consumption while still yielding materials with robust mechanical tunability) and cultured for 48 h under three different material conditions: 1) continuous flood exposure (λ = 470 nm, 1 mW cm−2) giving rise to softer gels; 2) dark culture yielding comparatively stiff materials; and 3) shuttered light exposure (1 min on, 4 min off), providing gels with cycled compliance (Figure 5b). The extent of softening (≈8% of initial moduli) and the dark material recovery rate (t1/2 ≈ 35 s) were similar for gels containing encapsulated cells in culture media (Figure 5b) or simply swollen in phosphate buffered saline (PBS) (Figure 3e,f), indicating that the system functions equally well in the presence or absence of complex biological components. High cell viability (≈95%) was observed in all conditions, demonstrating the overall cytocompatibility of gel formation and subsequent photomechanical material alterations (Figure 5c and Figure S5 (Supporting Information)). To determine the effects of each culture condition on fibroblast-to-myofibroblast differentiation, we quantified associated changes in luciferase activity of fibroblasts transfected with an αSMA–luciferase transcriptional reporter plasmid72 (Figure 5d). As expected, significant changes in αSMA transcriptional activity was observed for cells cultured between “stiff” and “soft” materials. Interestingly, myofibroblast activation was further enhanced within reversibly stiffening gels. This activating effect was reconfirmed by Western blot analysis of untransfected fibroblasts for expression of αSMA relative to housekeeper protein glyceraldehyde 3-phosphate dehydrogenase (Figure S6, Supporting Information). We further confirmed the observed effects using fibroblasts transfected with a Postn–luciferase transcriptional reporter plasmid,72 which acts as an additional transcriptional surrogate of myofibroblast differentiation. These findings are further substantiated by recent studies in 2D, where fibroblasts seeded on silicon membranes and subjected to cyclic stretching exhibited enhanced activation.73 Observed differences in 3D fibroblast transdifferentiation motivate further investigation, whereby differences in anisotropic cyclic mechanical loading are expected to play a large role in regulating powerful, yet largely unexplored, biological phenomena.
The results described here illustrate a generalizable strategy to create biomaterials whose mechanics can be cycled in response to user-defined inputs. Through the first demonstrated dual-chemoenzymatic modification of a recombinant protein, we create fusion species exhibiting stimuli-dependent intramolecular association that can be used as the basis for step-growth polymerization of gels. Implementation of the technique with the photoactive LOV2 protein yielded unprecedented reversible and spatiotemporal control over network dynamics, newly capturing critical aspects of the cellular microenvironment. These unique materials were used to investigate 3D biological response to cyclic loading, an important aspect of native ECM that has not been previously replicated in vitro. We expect that this approach will provide a new dimension to the growing field of mechanobiology, and will prove useful in the creation of a wide variety of protein-based and stimuli-responsive materials for tissue engineering, drug delivery, and many other applications.
Experimental Section
Synthesis and Purification of End-Functionalized Stimuli-Responsive Fusion Proteins: Plasmids encoding for CaMM13 and LOV2Jα, flanked with an N-terminal NMT- and a C-terminal sortase-recognition peptide sequence (MGNEASYPL and LPETG, respectively), were constructed using standard cloning techniques (Method S7, Supporting Information) and each cotransformed with those encoding for NMT/methionine aminopeptidase (Met-AP) into BL21(DE3) Escherichia coli (Thermo Fisher). Successful cotransformants were grown at 37 °C in lysogeny broth containing ampicillin (100 µg mL−1) and kanamycin (50 µg mL−1). After reaching an optical density of 0.6 (λ = 600 nm), isopropyl β-d-1-thiogalactopyranoside was added (final concentration of 0.5 × 10−3 m) to induce expression. Expression was carried out with or without 12-ADA (final concentration of 140 mg L−1, Method S5, Supporting Information) overnight under reduced temperature (18 °C). Cells were harvested via centrifugation and lysed by sonication. Clarified lysate was loaded onto HisPur Ni–NTA resin (Thermo Fisher), which was washed (20 × 10−3 m Tris, 50 × 10−3 m NaCl, 20 × 10−3 m imidazole) to remove unbound proteins. Following treatment of the resin with triglycine or an azide-containing polyglycine probe (20×, 4 h, 37 °C), sortagged proteins were eluted and purified by dialysis (molecular weight cutoff, MWCO ≈ 10 kDa). Protein identity and purity was confirmed by liquid chromatography–tandem mass spectrometry, SDS-PAGE, and gel shift analysis. Ultraviolet absorption (λ = 280 nm) and bicinchoninic acid assays were used to determine protein concentrations prior to use. Purified proteins remained stable for several weeks, based on their ability to form responsive gels, when kept refrigerated (4 °C) in PBS (pH = 7.4). Complete experimental details are given in Methods S7–S9 (Supporting Information).
Assessing Ca2+-Dependent Conformational Changes of CaMM13: Far-UV circular dichroism (CD) measurements were performed on a Jasco 720 CD spectrophotometer (1 mm path length, quartz cuvette) purged with nitrogen gas from 200 ≤λ ≤ 250 nm (scan rate = 100 nm min−1). Spectra were recorded for N3CaMM13N3 (25 μg mL−1) in buffer (20 × 10−3 m Tris, 30 × 10−3 m KCl) supplemented with either calcium (1 × 10−3 m CaCl2) or EGTA (1 × 10−3 m), and were normalized against protein-free controls.
Determining Extent of Protein Chemoenzymatic Modifications: To assess the ability for sortase and NMT to generate homogenous protein samples with quantitative reactivity, an SDS-PAGE gel shift assay was performed for each purified protein species (CaMM13, CaMM13N3, N3CaMM13, N3CaMM13N3, LOV2Jα, LOV2JαN3, N3LOV2Jα, N3LOV2JαN3). Following reaction (24 h at 37 °C) with mPEGBCN (100×) in buffer (20 × 10−3 m Tris, 50 × 10−3 m NaCl, 20 µL), denatured proteins (1 µg) were separated electrophoretically in polyacrylamide gels and visualized by Coomassie Brilliant Blue (Thermo Fisher) staining.
Step-Growth Polymerization of Protein–Polymer Hydrogels: Protein–polymer hydrogels were formed by SPAAC between PEGtetraBCN (2 × 10−3 m) and diazide crosslinking species in PBS (pH = 7.4). The diazide crosslinker mixture (4 × 10−3 m total) was comprised of modified fusion proteins (N3CaMM13N3 or N3LOV2JαN3, 0–3 × 10−3 m) and a balance of N3PEGN3 (1 × 10−3–4 × 10−3 m) to yield gels with 0, 25, 50, or 75 mol% protein crosslinker content. Upon component mixing, gelation was permitted to proceed for 2 h between Rain-X-treated glass slides (500 µm spacing) prior to slide separation and subsequent equilibration in PBS. For cell-laden gel formation, PEGtetraBCN was prereacted (1 h) with N3GRGDSNH2 peptide (0.5 × 10−3 m final in-gel concentration, Method S11, Supporting Information) prior to mixture with crosslinking species and cells in culture media. Protein–polymer gels based on CaMM13 and LOV2Jα each remained fully stable after two weeks of storage in cell culture media at 37 °C (Figure S7, Supporting Information), indicating that the PEGylated protein crosslinkers are not particularly prone to hydrolytic or enzymatic degradation.
Mechanical Testing of Protein–Polymer Gels: In situ rheological experiments were performed on optically thin (50 µm) protein–polymer gels formed between parallel quartz plates (8 mm diameter). The rheometer (Discovery HR-2, TA Instruments) was outfitted with a collimated blue light source (λ = 470 nm, 10 mW cm−2, ThorLabs) whose exposure was shuttered with an automated controller (NEARPOW) in experiments investigating the light-responsiveness of LOV2Jα gels (Figure S8, Supporting Information). The storage and loss moduli (G′ and G″, respectively) were measured by oscillatory rheology at constant strain (1%) and frequency (1 rad s−1), conditions determined to fall within the linear viscoelastic region of the gels. A thin coating of mineral oil around gel perimeter was employed to prevent evaporation during measurements.
Ca2+-Dependent Changes in CaMM13 Gel Swelling: CaMM13 gels (0–75 mol% protein content) were equilibrated in buffer (20 × 10−3 m 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 50 × 10−3 m NaCl), alternating between that supplemented with CaCl2 (1 × 10−3 m, 4 h) or EGTA (1 × 10−3 m, 8 h) to cycle gel swelling. Fractional mass changes, defined as the change in swollen gel mass relative to that of the initial swollen gel, were determined for each gel type after full equilibration and immediately preceding buffer exchange.
Spatiotemporal Control of Reversible Gel Softening: 50 mol% LOV2Jα gels were exposed to collimated blue light (λ = 470 nm, 1 mW cm−2, 30 s) through a patterned chrome photomask (Photo Sciences, Inc.). Gels were visualized before and immediately after light exposure through time-lapse videography using a digital camera (Nikon). Image analysis of gel color following flood light exposure (λ = 470 nm, 1 mW cm−2, 30 s) was performed (ImageJ) to determine yellow saturation content (interval = 5 s).
3T3 Fibroblast Cell Culture: Unmodified and transfected NIH 3T3 fibroblast cells were cultured in Dulbecco's modified Eagle's medium supplemented with fetal bovine serum (10%, Corning) and penicillin/streptomycin (1%, Corning) in a 5% CO2 atmosphere at 37 °C. Culture medium was changed every two to three days, and cells were passaged at ≈90% confluency. Following trypsinization, fibroblasts were encapsulated (5 × 106 cells mL−1) in 50 mol% LOV2Jα gels modified with N3GRGDSNH2 (0.5 × 10−3 m, Method S11, Supporting Information).
Cell Responses to Reversible Matrix Stiffness Modulation: NIH/3T3 fibroblasts were transfected with either an αSMA–luciferase or a Postn–luciferase promoter plasmid.72, 74 Transfection conditions (1.66 µg DNA µL−1 Lipofectamine 3000) were selected so as to maximize transfection efficiency (here, 65%) while retaining cell morphology and health. Transfected NIH/3T3 fibroblasts were encapsulated (5 × 106 cells mL−1) in 50 mol% LOV2Jα hydrogels in three polystyrene 24-well cell culture plates (Corning) under three different material conditions: 1) continuous flood exposure (λ = 470 nm, 1 mW cm−2) giving rise to softer gels; 2) dark culture yielding comparatively stiff materials; and 3) shuttered light exposure (1 min on, 4 min off). After cultured maintenance of these light exposure conditions for 48 h, samples were ground with a mortar and pestle and resuspended in lysis buffer (50 µL, 1% Triton X-100, 100 × 10−3 m Tris–HCl, 2 × 10−9 m ethylenediaminetetraacetic acid, 2 × 10−3 m dithiothreitol) prior to luciferase analysis (Pierce Firefly Luciferase Glow Assay, Thermo Fisher). Luciferase activity was measured in triplicate for each material condition.
Acknowledgements
The authors recognize and thank Dr. A. Nelson (University of Washington, UW) for generously providing access to their rheometer, Dr. L. Newman and Dr. R. Kahn (Emory University) for gifting the hNMT1 with Met-AP plasmid,75 as well as Dr. R. Warden-Rothman and Dr. A. Tsourkis (University of Pennsylvania) for providing the pSTEPL plasmid.76 Yellow Cameleon-Nano15/pBig was a gift from Takeharu Nagai (Addgene plasmid # 51962). pTriEx-PA-Rac1 was a gift from Klaus Hahn (Addgene plasmid # 22024). The authors thank B. Badeau for assistance in synthesizing N3OSu and FmocLys(N3)OH, as well as S. Adelmund for providing BCNOSu. The authors gratefully acknowledge support from S. Edgar at the UW Mass Spectrometry Center, the NIH and N. Peters at the UW W. M. Keck Microscopy Center (Grant No. S10 OD016240), as well as from D. Wise and N. Bragg (UW Marketing and Communications) for their assistance with visually analyzing spatiotemporal changes in LOV2Jα gel mechanics. This work was supported by a UW Faculty Startup Grant (C.A.D.), an Institute for Stem Cell & Regenerative Medicine Innovation Pilot Award (C.A.D.), a CAREER Award (DMR 1652141, C.A.D.) from the National Science Foundation, and a Pathway to Independence Award (Grant No. K99/R00 HL119353-01, J.D.) from the National Institutes of Health.
Conflict of Interest
The authors declare no conflict of interest.




