KIF5A p.Pro986Leu Risk Variant and Accelerated Progression of Amyotrophic Lateral Sclerosis
Funding: This work was supported by the Italian Ministry of Health (Grants R.F.-2021-12374238 to N.T. and R.F.-2018-12367768 to V.S. and A.R.). This research was supported by the Intramural Research Program of the National Institutes of Health (National Institute on Aging, project numbers 1ZIAAG000935, Bryan J. Traynor).
ABSTRACT
This study explored the impact of KIF5A rs113247976 (p.Pro986Leu), a risk allele for amyotrophic lateral sclerosis (ALS), on phenotypic variability in two Italian ALS cohorts (discovery, n = 865; replication, n = 1174). The minor allele (T) frequency was 0.015. No patients were homozygous (TT), allowing comparison between wild type and heterozygous carriers only. Heterozygous carriers showed faster disease progression (ALSFRS-R preslope). Findings were validated across both cohorts. Multiple linear regression identified p.Leu986 and age at onset as ALSFRS-R preslope predictors. In conclusion, heterozygous p.Leu986 in KIF5A is associated with faster ALS progression, supporting its consideration for genetic screening in clinical trials.
1 Introduction
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disorder affecting upper and lower motor neurons, resulting in progressive loss of independence due to muscular weakness and atrophy, and death often due to respiratory insufficiency. ALS is mainly sporadic, although in 5%–10% of cases a positive family history of motor neuron disease is reported, with high genetic heterogeneity [1].
In 2018, KIF5A, a member of the kinesin molecular motor superfamily expressed in neurons and involved in axonal transport, was identified as an ALS-associated gene [2]. In particular, the coding rs113247976 single nucleotide polymorphism (SNP) (p.Pro986Leu) was significantly associated with ALS risk, while an excess of loss-of-function mutations in the C-terminal cargo-binding domain was observed in ALS patients compared to controls [2]. While most KIF5A is typically cytosolic, unbound from cargo, and inhibited by head-tail association, ALS-associated mutations create a constitutively active kinesin lacking autoinhibition, leading to increased microtubule binding, altered dynamics, and accumulation in distal neurites, and altered axonal transport [3]. Furthermore, ALS-related KIF5A mutations alter KIF5A protein and RNA interactions, affecting RNA processing and cellular stress response pathways [3].
Interestingly, rare variants in other functional domains have already been associated with distinct neurodegenerative diseases (i.e., Hereditary Spastic Paraplegia type 10, Charcot–Marie-Tooth type 2, intractable myoclonus) [4, 5].
European ALS patients harboring KIF5A rare loss-of-function variants typically have a younger age at onset and prolonged survival [2], while very limited information is available for rs113247976. Our study aims to determine the frequency of the KIF5A rs113247976 in two Italian ALS cohorts and its impact on clinical variables associated with disease progression.
2 Methods
2.1 Participants and Clinical Assessment
We included 865 ALS Italian patients from the IRCCS Istituto Auxologico Italiano (discovery cohort) and 1174 from the Piemonte and Valle d'Aosta ALS Register (PARALS) (replication cohort) between 2013 and 2023 [6].
We recorded the following demographic and clinical data: sex, ALS family history, age at onset and at diagnosis, site of onset, survival after onset, and the ALSFRS-R preslope at the first evaluation (calculated as (48—ALSFRS-R score)/months between disease onset and evaluation). For the analysis of disease progression rate, we included only those patients for whom the first ALSFRS-R score available had been assessed within 45 days from diagnosis.
2.2 Genetic Analyses
For the discovery cohort, SNP genotyping data, obtained from previous Human 660 W-Quad BeadChips and Global Screening Arrays (Illumina) analyses [7], were used to impute the KIF5A rs113247976, as previously described [8]. For the replication cohort, whole-genome sequencing data were generated as previously reported [9]. All patients were screened for mutations in the main four ALS-associated genes (C9orf72, SOD1, TARDBP and FUS), as previously described [8].
2.3 Statistical Analyses
Statistical analyses were performed using RStudio version 2023.03. With no patients having the p.Leu986 homozygous genotype (TT), comparisons were made between the CC and CT genotypes. Cross-tabulated frequencies were calculated using Fisher's exact tests. The Wilcoxon-Mann–Whitney test was used for quantitative variables due to non-normality (Shapiro–Wilk test). Multiple linear regression explored the impact of several variables on ALSFRS-R preslope. The rs113247976 genotype's effect on survival was estimated using Kaplan–Meier and Cox regression analyses. Censoring was applied for patients alive at last follow-up, and missing data were handled by pairwise deletion.
3 Results
Demographic and clinical features, and genotype data for the coding KIF5A rs113247976 (p.Pro986Leu) of the discovery (n = 875 patients) and replication (n = 1174 patients) Italian ALS cohorts are reported in Table 1. Compared to the discovery cohort, the replication one had a higher median age at onset by up to 6 years, longer survival by 6–7 years, more bulbar cases, more female patients, and a higher prevalence of familial cases. Additionally, more patients carried mutations in the four main ALS genes. No patients with the p.Leu986 variant in homozygous state were found, and both the ALSFRS-R preslope and the minor allele frequency (MAF) of the p.Leu986 variant did not significantly differ between the two cohorts (Table 1). Further, rs113247976 MAF was similar (0.014 and 0.017) to previously described ALS populations from various European countries and the United States (0.02) [2]. For the replication cohort, additional rare (MAF < 0.1%) variants in minor ALS-related genes and their frequency in p.Pro986 and p.Leu986 carriers are reported in Supporting Information S1 and Table S1.
| Variable | Discovery cohort | Replication cohort | p | ||
|---|---|---|---|---|---|
| No patients (frequency) | Median (IQR) | No patients (frequency) | Median (IQR) | ||
| Sex | 865 | 1174 | 1.18e-05 | ||
| Male | 554 (64.0%) | 638 (54.3%) | |||
| Female | 311 (36.0%) | 536 (45.7%) | |||
| ALS family history | 833 | 1170 | 3.81e-07 | ||
| FALS | 20 (2.4%) | 113 (9.6%) | |||
| SALS | 813 (97.6%) | 1057 (90.0%) | |||
| Mutations in common ALS genes | 865 | 1174 | 5.40e-06 | ||
| C9orf72 | 20 (2.3%) | 80 (6.8%) | |||
| SOD1 | 0 (0.0%) | 25 (0.2%) | |||
| TARDBP | 7 (0.8%) | 16 (1.4%) | |||
| FUS | 2 (0.2%) | 6 (0.5%) | |||
| Age at onset (years) | 798 | 61.6 (52.7–69.6) | 1174 | 67.7 (59.7–74.1) | 1.93e-26 |
| Survival from onset (months) | 776 | 25.3 (13.1–46.1) | 1174 | 32.7 (20.7–61.8) | 4.37e-15 |
| Site of onset | 865 | 1174 | 8.55e-05 | ||
| Bulbar | 206 (23.8%) | 374 (31.9%) | |||
| Spinal | 659 (76.2%) | 800 (68.1%) | |||
| ALSFRS-R preslopea | 449 | 0.60 (0.32–1.04) | 1174 | 0.59 (0.29–1.16) | 0.54 |
| rs113247976 SNP genotype | 865 | 1174 | 0.56 | ||
| p.Pro986/p.Pro986 | 841 (97.2%) | 1135 (96.7%) | |||
| p.Pro986/p.Leu986 | 24 (2.8%) | 39 (3.3%) | |||
| MAF p.Leu986 | 0.014 | 0.017 | 0.57 | ||
- Abbreviations: ALS, amyotrophic lateral sclerosis; ALSFRS-R, ALS Functional Rating Scale Revised; FALS, familial ALS; IQR, interquartile range; MAF, minor allele frequency; no, number; SALS, sporadic ALS; SNP, single nucleotide polymorphism.
- a Only patients for whom the ALSFRS-R score was assessed < 45 days from diagnosis were included. Significant p values (< 0.05) are reported in bold.
In both cohorts, heterozygous p.Leu986 carriers showed a higher ALSFRS-R preslope than homozygous p.Pro986 patients [discovery: 1.25 (IQR 0.65–2.42) vs. 0.59 (IQR 0.31–1.01), p = 0.0025; replication: 0.90 (IQR 0.34–1.98) vs. 0.59 (IQR 0.29–1.14), p = 0.036] (Figure 1A,B). Given the similar median values across cohorts, a combined analysis was performed, confirming this association [1.09 (IQR 0.37–2.06) vs. 0.59 (IQR 0.29–1.10), p = 0.00084] (Figure 1C).
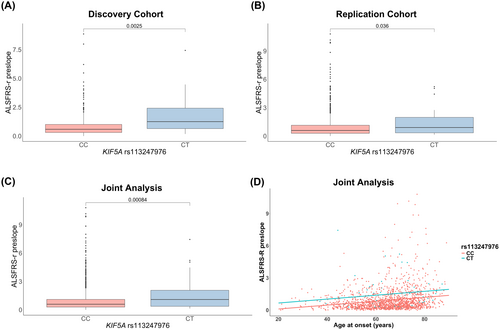
Multiple linear regression showed that only the rs113247976 KIF5A genotype and age at onset significantly predicted the ALSFRS-R preslope (p = 0.000246 and 3.51e-14), unlike gender, onset site, and C9orf72 status. After refining the model, it continued to effectively predict the ALSFRS-R preslope [F(2, 1620) = 35.48; p = 8.323e-16], despite explaining limited variance (adjusted R2 = 0.04078) (Figure 1D; Table S2).
Conversely, we found no significant differences in the other demographic and clinical features collected, including survival after onset (Figure S1 and Table S3), between the p.Pro986/p.Pro986 and p.Pro986/p.Leu986 genotypes in both the discovery and replication cohorts, and their joint analysis.
4 Discussion
Our data support the involvement of the common KIF5A p.Leu986 variant, a known ALS risk factor, in influencing disease progression. Independently validated across discovery, replication, and combined cohorts, our findings show that p.Leu986 carriers experience faster early disease progression, as indicated by the ALSFRS-R preslope. However, this early decline does not correlate with reduced survival, contradicting prior, statistically non-significant suggestions of shorter disease duration in p.Leu986 carriers [5].
Noteworthy, among functional and disability scores, the ALSFRS-R, which is widely recognized for its multidimensional approach to measuring functional impairment in ALS (i.e., bulbar, fine and gross motor domains, and respiratory function), has been shown to be significantly related to ALS outcome and prognosis [10].
Although the ALSFRS-R score reflects functional decline, it does not consistently predict survival, as patients with the same score may have different affected domains, which can impact survival differently. This variability may partly explain the absence of survival differences despite faster disease progression associated with the p.Leu986 variant [11].
ALS progression is known to be influenced by a complex interplay of genetic and non-genetic factors (i.e., age at onset, site of onset, psychosocial factors, body mass index) [12]. Regarding genetic determinants, repeat expansions in C9orf72, mutations in FUS, and intermediate-length CAG repeats in ATXN2 have been linked to faster disease progression [13, 14]. Conversely, individuals with either double mutations or the single p.D90A mutation in SOD1, as well as young patients with ALS2 and SETX-related autosomal recessive ALS, tend to experience a slower disease progression [13]. In addition, ATXN2 patients exhibit more frequent spinal onset, concurrent frontotemporal dementia, and shorter survival up to 1 year [14]. Except for the UNC13A rs12608932 and the CAMTA1 rs2412208 risk alleles, which influence shorter survival [8, 15], common variants have typically limited impact on progression outcomes (i.e., survival, age at onset). Notably, the simultaneous presence of multiple risk alleles (i.e., UNC13A rs12608932, CAMTA1 rs2412208) or repeat expansions (i.e., C9orf72, ATXN2) is associated with significantly reduced survival [16]. Within this framework, our study underscores the importance of the KIF5A rs113247976 variant as a predictor of accelerated disease progression in early stages, thereby contributing valuable insights into the genetic modifiers of ALS.
However, a limitation of our study is that the discovery cohort comprises only Italian ALS patients from a single center, unlike the replication cohort, which comes from a prospective, population-based registry in Northern Italy. This distinction accounts for the observed epidemiological and clinical discrepancies between the two cohorts. Nonetheless, the ALSFRS-R preslope, measured within 45 days of diagnosis, and the MAF of the p.Leu986 variant were comparable across both cohorts, facilitating their comparative analysis. Moreover, the ALSFRS-R score tends to decline curvilinearly rather than linearly, contradicting the assumption of a uniform progression rate [17]. This non-linear pattern suggests that the initial ALSFRS-R preslope might differ based on the time elapsed since onset. To mitigate potential bias introduced by assessing ALSFRS-R scores at different disease durations, we focused our analysis on patients whose ALSFRS-R evaluations were conducted within 45 days of diagnosis, in order to standardize the effect of disease duration on our results.
Overall, this research provides significant insights into the limited body of knowledge concerning the role of KIF5A in ALS progression. As ALS is highly heterogeneous, the elucidation of genetic and environmental determinants that influence its course is crucial for anticipating the potential success of therapeutic interventions. In different clinical trials, indeed, only a subset of patients responded to treatments, underscoring the need for patient stratification in future studies [18]. In this scenario, preliminary screening for variants associated with phenotypic traits in ALS, such as KIF5A rs113247976, which in our study is linked to an accelerated progression, could prove vital in forecasting the outcomes of treatment strategies and informing the selection of participants for forthcoming clinical trials and could enhance the power of the ENCALS Survival Prediction Model.
Author Contributions
Arianna Manini, Alberto Brusati, Antonia Ratti and Nicola Ticozzi: conceptualization. Arianna Manini, Rosario Vasta, Alberto Brusati, Davide Gentilini and Nicola Ticozzi: methodology. Arianna Manini, Rosario Vasta and Alberto Brusati: formal analysis. Arianna Manini, Rosario Vasta, Alessio Maranzano, Francesco Gentile, Stefano Messina, Federico Verde, Cristina Moglia and Nicola Ticozzi: investigation. Antonia Ratti, Vincenzo Silani, Adriano Chiò, John E. Landers, Bryan J. Traynor, Adriano Chiò and Nicola Ticozzi: resources. Arianna Manini, Rosario Vasta, Alberto Brusati, Alessio Maranzano, Francesco Gentile, Eleonora Colombo and Nicola Ticozzi: data curation. Arianna Manini: writing – original draft preparation. Arianna Manini, Rosario Vasta, Antonia Ratti and Nicola Ticozzi: writing – review and editing. Antonia Ratti, Vincenzo Silani and Nicola Ticozzi: supervision. Antonia Ratti and Nicola Ticozzi: project administration. Vincenzo Silani, Antonia Ratti and Nicola Ticozzi: funding acquisition. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors thank all the patients who participated in this study and their caregivers. This work was financially supported by the Italian Ministry of Health (Grants RF-2021-12374238 to N.T. and RF-2018-12367768 to V.S. and A.R.). This research was supported by the Intramural Research Program of the National Institutes of Health (National Institute on Aging, project numbers 1ZIAAG000935, Bryan J. Traynor). The funders had no role in data collection or analysis and did not participate in writing or approving the manuscript. A.R. acknowledges “Aldo Ravelli Center for Neurotechnology and Experimental Brain Therapeutics”, Università degli Studi di Milano. The Authors acknowledge the Italian Ministry of Health [“Piano Nazionale Complementare Ecosistema Innovativo della Salute—Hub Life Science-Diagnostica Avanzata (HLS-DA)”—PNC-E3-2022-23683266—“INNOVA”], the Italian Ministry of Education and Research—MUR (“Dipartimenti di Eccellenza” Program 2023–2027—Dept. of Pathophysiology and Transplantation, Università degli Studi di Milano) and the ERN Euro-NMD for support. Open access publishing facilitated by Universita degli Studi di Milano, as part of the Wiley - CRUI-CARE agreement.
Ethics Statement
The study was approved by the Ethics Committee of the IRCCS Instituto Auxologico Italiano (2021_05_18) and by the Ethics Committee of the Turin ALS Center (Comitato Etico Azienda Ospedaliero-Universitaria Città della Salute e della Scienza, Torino). Written informed consent was obtained from each patient at the time of evaluation to use semi-anonymized clinical data for research purposes. The study was conducted in accordance with the principles of the Declaration of Helsinki.
Conflicts of Interest
V.S. received compensation for consulting services and/or speaking activities from AveXis, Cytokinetics, Italfarmaco, Liquidweb Srl, Novartis Pharma AG, and Zambon Biotech SA. He is on the Editorial Board of Amyotrophic Lateral Sclerosis and Frontotemporal Degeneration, European Neurology, American Journal of Neurodegenerative Diseases, Frontiers in Neurology, and Exploration of Neuroprotective Therapy. N.T. received compensation for consulting services from Amylyx Pharmaceuticals, Biogen, Italfarmaco, and Zambon Biotech SA. He is Associate Editor for Frontiers in Aging Neuroscience.
Open Research
Data Availability Statement
Anonymized raw data are archived on Zenodo (doi: 10.5281/zenodo.10953602) and are available upon request to the corresponding author.




