Neuronal intranuclear inclusion disease is genetically heterogeneous
Abstract
Neuronal intranuclear inclusion disease (NIID) is a clinically heterogeneous neurodegenerative condition characterized by pathological intranuclear eosinophilic inclusions. A CGG repeat expansion in NOTCH2NLC was recently identified to be associated with NIID in patients of Japanese descent. We screened pathologically confirmed European NIID, cases of neurodegenerative disease with intranuclear inclusions and applied in silico-based screening using whole-genome sequencing data from 20 536 participants in the 100 000 Genomes Project. We identified a single European case harbouring the pathogenic repeat expansion with a distinct haplotype structure. Thus, we propose new diagnostic criteria as European NIID represents a distinct disease entity from East Asian cases.
Introduction
Neuronal intranuclear inclusion disease (NIID) is a clinically heterogeneous, multi-system neurodegenerative condition with manifestations comprising cognitive impairment, parkinsonism, and neuropathy at varying ages of onset.1 Central to the pathological diagnosis is presence of characteristic intranuclear eosinophilic ubiquitinated inclusions in both neuronal and non-neuronal cells. Despite the first case being described in 1968,2 a CGG repeat expansion in NOTCH2NLC has only been found recently to be associated with NIID in Japanese patients.3, 4 This was prompted by the clinico-pathological overlap with Fragile X-associated tremor-ataxia syndrome (FXTAS)5 and increasing recognition of noncoding repeat expansions being crucially causative in neurological disorders.3, 4 Since these findings, the same expansion has been reported in several East Asian cohorts including Chinese patients with skin-biopsy proven NIID6, 7; Chinese essential tremor cases8 and Japanese leukodystrophy cases.9
Inspired by the high prevalence of this expansion within East Asian patients, we instigated screening for the repeat within Europeans with pathological confirmation of neuronal and/or glial hyaline intranuclear inclusions on brain tissue to further understand the molecular basis of disease. The very similar intranuclear inclusions seen in NIID can occur concomitantly with another proteinopathy. Therefore, we also screened post-mortem cases with neuronal intranuclear inclusions (NIIs) in other neurodegenerative diseases with the aim of assessing whether clinically heterogeneous presentations converge on a common proteinopathy aggregate. Lastly, we applied in silico-based screening of a deeply-characterized cohort of 20,536 patients with neurological presentations enrolled in the 100,000 Genomes Project to characterize the prevalence within a predominantly European population.10 We show that the NOTCH2NLC repeat expansion is a rare cause of NIID in Europeans and that at least two distinct disease entities exist under the name NIID.
Methods
Case selection
The study was approved by UCL Institute of Neurology Institutional Review Board. Tissue and DNA samples from other institutions met approval from local ethics boards. Eleven NIID cases (Cases 1–11) were identified from: Queen Square Brain Bank (QSBB)11; Spain (IDIBAPS Brain Bank Barcelona)5; Finland12; Australia (South Australian Brain Bank13 and Macquarie University) and USA (Mayo Clinic). Thirteen cases with primary protein misfolding pathology and NIIs (Cases 2-1 to 2-13) were included from: QSBB; Austria (Vienna Brain Bank)14; IDIBAPS5 and Mayo Clinic. Five cases of FTLD-FUS were also included from IDIBAPS and QSBB (Cases 2-14 to 2-18). We used a Japanese patient previously described with NOTCH2NLC repeat expansion-associated NIID (Case J) as a positive control.3 DNA extraction from QSBB, Spain, and USA samples of fresh frozen cerebellar tissue was carried out as per Qiagen Gentra Puregene Tissue Kit protocol (concentration ≥ 219.7 ng/µL).
Repeat-primed polymerase chain reaction and fragment analysis
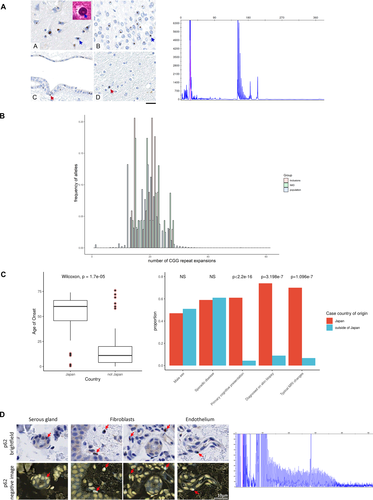
Repeat-primed polymerase chain reaction (RP-PCR) was designed as described3 to assess for CGG repeat expansion using genomic DNA. RP-PCR analysis was performed using primers: 5’-AGCGCCCACAGCAGAGCGGC-3’; 5’-CCGGGAGCTGCATGTGTCAGAGGCGGCGGCGGCGGCGG-3’; 5’-(FAM)-CCGGGAGCTGCATGTGTCAGAGG3’, LA taq with GC buffer (TaKaRa Bio) and deaza-dGTP. The PCR protocol used initial denaturation at 95°C for 5 minutes, followed by 50 cycles of 95°C for 30 seconds, 98°C for 10s, 62°C for 30 seconds and 72°C for 2 minutes. The ramp rate to 95°C and 72°C was 2.5°C per second and that to 62°C was 1.5°C per second3. For fragment analysis, 9.2 µL HiDi formamide was combined with 0.5 µL LIZ 500 size standard per 1 µL PCR product. FAM-labeled PCR products were denatured at 95 °C for three minutes and on ice for three minutes then separated on ABI3730 DNA Analyser (ThermoFisher). Electropherograms were visualized on GENEMAPPER (ThermoFisher). We judged a sawtooth tail pattern in the electropherogram as the disease-associated repeat expansion (Figure 1). This process was replicated three times, with three positive controls to ensure negative results did not arise from technical error. Estimating repeat size from fragment analysis employed previously described protocol.3
Whole-genome sequence analysis for repeat expansion
We used ExpansionHunter v.2,15, 16 a validated tool that identifies repeat expansions using whole-genome sequencing (WGS) data. We searched for “CGG” repeats within the genomic co-ordinates of the repeat expansion (Chr1:149390802-149390841, GRCh38) in a cohort of 20,536 patients with neurological presentation recruited into the 100 000 Genomes Project.10 Interruptions within the repeat sequence were accounted for in the algorithm. Ethnicities were estimated using a random forest classifier based on 1,000 Genomes Project as a training dataset.
Genotyping
Sample processing for Illumina GSAv2.0 arrays was carried out according to Infinium HTS Assay protocol (Illumina Inc.) at UCL Genomics. Three hundred nanogram of DNA was whole-genome amplified, fragmented, precipitated, and resuspended in hybridization buffer. Samples were hybridized onto Illumina GSA beadchips and incubated at 48 °C for 16 hours. Beadchips were stained then scanned using iScan (Illumina). Total genotyping rate was 0.993. Principal components were calculated using PLINK v.1.917 and population stratification analysis for inferred ancestries using Peddy (Python).
Haplotype analysis
Haplotype blocks were estimated based on 90% confidence intervals of D’ disequilibrium statistic for pairs of variants (PLINK17). The haplotype analysis was set within the NOTCH2NL paralogous region (Chr1:120705588–149410843, GRCh38) containing 380 genotyped SNPs. The genotyped SNP overlap between the three patient groups compared (NOTCH2NLC expansion-negative European NIID, Case 12 and Case J) was high at 96.7% remaining consistent at 96.3% with minor allele frequency (MAF) >0.05.
Comparisons of clinical characteristics
We reviewed Medline and Pubmed databases for cases of “neuronal intranuclear inclusion disease”; “neuronal intranuclear hyaline disease”; “neuronal intranuclear hyaline inclusion disease” and “intranuclear hyaline inclusion disease,” using key search terms as applied, without a date restriction. We identified 145 independent cases of NIID reported in the literature (April 2019). All statistical analyses were executed in R (version 3.5.1).
Results
NIID is genetically and phenotypically heterogeneous
We find no evidence of the repeat to a pathological level within eleven NIID cases of European ancestry confirmed on post-mortem brain examination (Table 1: Cases 1–11). These cases have been well-characterized including a monozygotic twin with juvenile-onset movement disorder, from whom the term NIID was coined12; as well as other cases with both juvenile-onset11 and adult-onset5, 13 disease. Revisiting the pathology confirmed that NIIs stained positive for p6211 further validating the diagnosis (Figure 1A). The median number of CGG repeats in NOTCH2NLC was 20 (range 14–28) in these patients (Figure 1B), falling within the range of repeats seen in asymptomatic Far East populations.3, 4 This suggests genetically heterogeneous mechanisms underlie NIID in European patients. In support of this diverse underlying molecular mechanism is the dichotomy in clinical presentation between non-Japanese and Japanese NIID cases. Of 145 reported NIID cases, two thirds are from Japan (100 cases) and are of an older age of onset compared to non-Japanese cases (median (IQR): 60 years (46–66) and 11 years (4–20) respectively, Wilcoxon rank sum P-value = 1.67e-5). Most Japanese patients had a primary cognitive presentation (61%), with a large proportion of cases having pathognomonic MRI changes at the corticomedullary junction (70%). Furthermore, 74% of Japanese cases were diagnosed on antemortem skin biopsy compared with ~ 9% of non-Japanese cases reflecting the lack of extraneuronal involvement in cases outside of Japan12 (Figure 1C). Deeper comparison of the inclusions has demonstrated differences also in their composition; inclusions were likely filamentous in European cases12 without the fine granular material reported in Japanese cases.1
| Case ID | Estimated number of CGG repeats | Age of onset | Sex | Family history | Country of origin | Clinical Diagnosis/ Presentation pre-biopsy | Main pathological findings and diagnosis | Other pathological findings | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Allele 1 | Allele 2 | |||||||||
| Pathologically-confirmed NIID | 11 | 21 | - | 17 | M | Yes | UK | Parkinsonism, tremor, bulbar and autonomic symptoms. Died aged 24 years. | NIID: widespread neuronal hyaline intranuclear inclusions immunoreactive for ubiquitin and p62 | See Figure 1C |
| 22 | 22 | 28 | 33 | M | Yes | Australia | Slowly progressive motor and sensory neuronopathy with ataxia. Death at 46 years. | NIID: eosinophilic neuronal intranuclear inclusions | Degeneration of substantia nigra, medial thalamus and cerebellum | |
| 32 | 15 | 20 | 60s | F | No | Australia | Unknown presentation. Death aged 67 years. | NIID: cortical neurons especially large pyramidal cells show eosinophilic intranuclear inclusions | No overt neuronal loss from the cerebral cortex and no reactive astrogliosis | |
| 4 | 15 | 23 | 52 | F | No | Australia | Slowly progressive primary lateral sclerosis. Death aged 72 years. | NIID: neuronal and astrocytic intranuclear inclusions throughout the cerebral cortex | Upper motor neuron loss and lateral corticospinal tract degeneration | |
| 53 | 19 | 22 | 11 | F | Yes (MZ twin) | Finland | Ataxia, rage, seizures and extrapyramidal symptoms. Death aged 21years. | NIID: inclusion bodies in most nerve cell types of central and peripheral nervous systems | Inclusions also seen in the retina and subtotal loss of nigral neurons | |
| 64 | 15 | 25 | 49 | F | Yes | Spain | Ataxia. Death aged 62 years. | NIID: abundant glial nuclear inclusions | Rosai-Dorfman disease (Case 3 Gelpi et al.) | |
| 74 | 16 | 23 | 82 | F | Yes | Spain | Dementia. Death aged 84 years. | NIID: abundant glial nuclear inclusions | ARTAG and SVD (Case 2 Gelpi et al.) | |
| 8 | 17 | 23 | 26 | F | No | USA | Clinical diagnosis unclear | NIID | ||
| 9 | 15 | 19 | 84 | M | No | USA | Alzheimer’s disease, ataxia | NIID: intranuclear hyaline inclusions in neurons and glia in widespread areas of the brain | Hippocampal sclerosis, argyrophilic grain disease, Braak 0, Thal 1, TDP 1 | |
| 10 | 14 | 27 | 69 | M | No | USA | Diagnosed clinically with NIID | NIID: neuronal intranuclear inclusions | ||
| 11 | 19 | - | 80 | M | No | USA | Unknown presentation | NIID | Inferior olivary hypertrophy | |
| 12 | 19 | expanded | 51 | F | No | Ukraine | Relapsing encephalopathy and migraines | Antemortem skin biopsy contains p62 positive intranuclear inclusions | ||
| Case ID | Estimated number of CGG repeats | Age of onset | Sex | Family history | Country of origin | Clinical Diagnosis/ Presentation pre-biopsy | Main pathology | Presence of p62/ ubiquitin-positive and TDP43 or FUS-negative NIIs | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Allele 1 | Allele 2 | |||||||||
| Other pathology with neuronal intranuclear inclusions (NIIs) | 2-1 | 15 | 19 | 49 | F | Unknown | UK | Parkinson’s disease | Parkinson’s disease, Braak 6 (diffuse neocortical) | Medial temporal lobe |
| 2-2 | 15 | 23 | 49 | M | Yes (PD in distant family) | UK | Progressive supranuclear palsy | Parkinson’s disease, Braak 4 (limbic) | Medial temporal lobe | |
| 2-3 | 15 | 22 | 51 | F | No | UK | Corticobasal syndrome | Alzheimer’s disease with amygdala-restricted Lewy pathology | Medial temporal lobe | |
| 2-4 | 15 | 21 | 56 | M | Unknown | UK | Progressive supranuclear palsy | Progressive supranuclear palsy | Medial temporal lobe and neocortex | |
| 2-5 | 20 | - | 78 | F | Unknown | UK | Corticobasal syndrome | Corticobasal degeneration | Medial temporal lobe, neocortex and cerebellum | |
| 2-6 | 14 | 22 | 60 | M | No | UK | Atypical progressive aphasia | Intermediate level Alzheimer’s disease pathological change | Medial temporal lobe | |
| 2-7 | 15 | 22 | 83 | M | No | UK | Parkinson’s disease | Parkinson’s disease, Braak 5 (limbic) | Medial temporal lobe | |
| 2-8 | 18 | 27 | 64 | M | No | UK | Corticobasal syndrome | Lewy pathology, Braak 5 (limbic); Cerebellar degeneration of unknown nature | Medial temporal lobe, neocortex and cerebellum | |
| 2-9 | 18 | 27 | 66 | F | No | USA | Dementia | Dementia with Lewy bodies | Present | |
| 2-104 | 22 | 23 | 47 | M | Yes | Spain | Rapidly progressive ataxia/ parkinsonism | Fatal familiaI insomnia and incidental inclusions (Case 4 in Gelpi et al. 2017) | Hippocampus | |
| 2-11 | 21 | 22 | 77 | M | Unknown | Spain | FXTAS | FXTAS, NFT II, AGD I and SVD | Present | |
| 2-12 | 21 | 28 | 86 | N | Unknown | Austria | Severe dementia | Vascular encephalopathy, AGD, ARTAG, AD A2B2C2, CAA type 1 + 2 | Present | |
| 2-135 | 15 | 20 | 63 | F | Unknown | Austria | Rapidly progressive dementia, pyramidal and extrapyramidal symptoms | MM1/MV1 CJD, Braak and Braak stage II | Cerebellar Purkinje cells | |
| 2-14 | 21 | - | 56 | M | Unknown | Spain | Fronto-temporal dementia | FTLD-ALS-FUS (ubiquitin), LBD (olfactory bulb) | None | |
| 2-15 | 19 | 23 | 69 | F | No | UK | Fronto-temporal dementia | FTLD-FUS | None (NIFID) | |
| 2-16 | 16 | 21 | 44 | M | Unknown | UK | Fronto-temporal dementia | FTLD-FUS | None (NIFID) | |
| 2-17 | 20 | 21 | 41 | F | No | UK | Fronto-temporal dementia | FTLD-FUS | None (NIFID) | |
| 2-18 | 14 | 22 | 49 | M | No | UK | Fronto-temporal dementia | FTLD-FUS | None (FTLD-U) | |
- Estimated number of CGG repeats using fragment analysis in our patients with NIID (Cases 1 to 12) and in other cases with concomitant intranuclear inclusions and with inclusions associated to other proteinopathies (Cases 2-1 to 2-13 and cases of FTLD-FUS: Cases 2-14 to 2-18). Where the sizing is not applicable (-), it is likely that the allele may be homozygous for the number of repeats in that patient providing overlapping traces and this allele is not expanded as no sawtooth pattern is visualized in comparison to our positive control. ABC score: A, amyloid phase according to Thal; B, Braak and Braak neurofibrillary stage; C, neuritic plaque score according to CERAD (each score ranges from 0 to 3); AD, Alzheimer's disease neuropathological changes; AGD, argyrophilic grain disease; ARTAG, aging-related tau astrogliopathy; CAA, cerebral amyloid angiopathy; CJD, Creutzfeldt-Jakob disease; FTLD, frontotemporal dementia; FTLD-FUS, FTLD-fused in sarcoma subtype; FTLD-ALS-FUS, FTLD and amyotrophic lateral sclerosis of FUS-subtype; FXTAS, fragile X-associated tremor/ataxia syndrome; LBD, Lewy body disease; MM, methionine homozygosity at codon 129 of the PRNP gene; MV, methionine valine heterozygous genotype at codon 129 of the PRNP gene; MZ twin, monozygotic twin; NIFID, neuronal intermediate filament inclusion disease; NIID, neuronal intraneuronal inclusion disease; NIIs, neuronal intranuclear inclusions; NFT, neurofibrillary tangles; PD, Parkinson’s disease; SVD, small vessel disease.
NOTCH2NLC repeat expansion does not underlie other neurodegenerative diseases with secondary intranuclear inclusions
Further confounding the diagnostic definition of NIID is the presence of similar intranuclear inclusions with concomitant protein-misfolding pathology.5 FXTAS was excluded from these cases. To investigate the underlying pathophysiology of such disorders, we screened a cohort of 13 cases with primary pathology in addition to NIIs (Table 1: Cases 2-1 to 2-13, Supplementary Figure S1). Within QSBB, ten cases were found to have intranuclear inclusions with positive staining for p62 and ubiquitin out of 850 brain samples. The other cases have been previously reported5, 14 in a range of presentations such as with coexisting prion disease.14 We further screened specific cases of FTLD-FUS subtype (Table 1: Cases 2-14 to 2-18) where the alike intranuclear inclusions have FUS recruited within. We also found no evidence of the repeat expansion within this cohort, which harbour similar estimated CGG repeats as seen in the asymptomatic population (median 20.5, IQR 16–22) (Figure 1B). This suggests that the abnormal repeat expansion in NOTCH2NLC is not the only driver for diseases with NIIs and highlights that multiple pathways are likely to converge on the end-product of intranuclear inclusion formation.
Frequency of repeat expansion within the European population
We have shown that the repeat expansion is not found in any of our European patients with pathologically-proven NIID compared to pathogenic expansions in 93–100% of Japanese and Chinese patients.3, 4 Leveraging the availability of WGS data in a large cohort of 20 536 deeply phenotyped participants presenting with neurological symptoms recruited in the 100 000 Genomes Project,10 we found the median number of NOTCH2NLC CGG repeats to be 20 (IQR 16–22) within this population (Figure 1B). The number of repeat expansions in our cohort of NIID patients and in those with pathological intranuclear inclusions did not differ significantly from this ‘background’ population (ANOVA p> 0.05). Furthermore, there were no significant differences in the number of repeats among ethnic groups (Supplementary Figure S2). Fragment analysis was used to verify the expansion size in ten individuals who had an estimated repeat size greater than 40 on one allele as ascertained using ExpansionHunter. In a patient with 58 repeats on one allele estimated from ExpansionHunter, fragment analysis demonstrated a pathogenic repeat expansion in a 59-year-old woman of Ukrainian ancestry who presented with a 10-year history of recurrent encephalopathy and migraines (Case 12). The patient was reviewed with respect to these results and subsequent skin biopsy revealed intranuclear p62 and ubiquitin-positive inclusions, confirming a diagnosis of NIID (Figure 1D).
Prompted by our observation of the low prevalence (approximately 1 in 20,000) of the pathogenic repeat expansion within a European population and lack of expansion within pathologically-confirmed cases, analyses of principal components and inferred genetic ancestry showed that the Ukrainian patient (Case 12) had no overlapping ancestry with the Japanese patient (Case J) (Supplementary Figure S3). Analysis of the entire NOTCH2 region encompassing associated paralogs revealed 27 haplotype blocks from the genotyped SNPs although no SNPs overlapped with expansion-containing region. This showed differing haplotypes for the Ukrainian patient (Case 12), European NOTCH2NLC-CGG-negative NIID patients (Cases 1–11) and the expansion-positive Japanese patient (Case J), even for SNPs with MAF>0.05 (Supplementary Table S1). Thus, presence of the rare repeat expansion in our European patient has likely arisen from a separate founder effect to that seen in Japanese cases.
Discussion
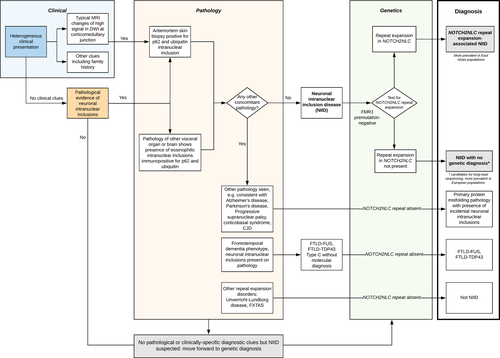
These results suggest that European NIID cases arise through a separate pathophysiological process to East Asian patients despite both diseases converging on the same signature of abnormal intranuclear inclusions. These differences in genetic, clinical, and pathological features suggest that at least two distinct disease entities exist under the name NIID. While Far East cases are driven by repeat expansion in NOTCH2NLC, in the single patient of European ancestry diagnosed with NIID due to NOTCH2NLC repeat expansion, haplotype analysis suggested a separate, rarer, founder mutation than that in Japanese cases. Further characterization of the genetic associations with NIID in other populations would be important although we are limited in the number of cases available. We therefore propose new criteria for characterization of NII-associated disorders (Figure 2) distinguishing between diseases with primary and secondary NIIs partitioned by pathological and molecular features. Thus, our findings are important by showing that the NOTCH2NLC repeat expansion is not the only cause underlying NIID pathogenesis or NII formation.
Acknowledgments
The authors thank the participants and their families for their help with this work. We are grateful to Professor Shoji Tsuji, University of Tokyo, for his contribution of the positive control case from Japan for our study. The authors thank Professor Janice Holton from the Queen Square Brain Bank for reporting and identifying cases used for this paper. The authors thank the Neurological Tissue Bank of the Biobank-Hospital Clinic – IDIBAPS, Barcelona, Spain and Teresa Ximelis for sample and data procurement. We are grateful to Dr Rebecca Ormsby and the South Australian Brank Bank for providing samples. ZC was funded by a Leonard Wolfson Clinical Research Fellowship (Grant number 157793). ZJ is supported by the Department of Health’s NIHR UCLH/UCL Biomedical Research Centre’s funding scheme. AT is a Medical Research Council Clinician Scientist (MR/S006753/1). TL is supported by an Alzheimer’s Research UK Senior Fellowship. We would also like to thank the MSA Trust, Medical Research Council (MR/S01165X/1), Wellcome Trust (WT093205 MA and WT104033AIA), Ataxia UK and Rosetrees Trust for funding. This research was made possible through access to the data and findings generated by the 100,000 Genomes Project. The 100,000 Genomes Project is managed by Genomics England Limited (a wholly owned company of the Department of Health and Social Care). The 100,000 Genomes Project is funded by the National Institute for Health Research and NHS England. The Wellcome Trust, Cancer Research UK and the Medical Research Council have also funded research infrastructure. The 100,000 Genomes Project uses data provided by patients and collected by the National Health Service as part of their care and support.
Genomics England Research Consortium
Ambrose J. C.1, Arumugam P.1, Baple E. L.1, Bleda M.1, Boardman-Pretty F.1,2, Boissiere J. M.1, Boustred C. R.1, Brittain H.1, Caulfield M. J.1,2, Chan G. C.1, Craig C. E. H.1, Daugherty L. C.1, de Burca A.1, Devereau A.1, Elgar G.1,2, Foulger R. E.1, Fowler T.1, Furió-Tarí P.1, Hackett J. M.1, Halai D.1, Hamblin A.1, Henderson S.1,2, Holman J. E.1, Hubbard T. J. P.1, Ibáñez K.1,2, Jackson R.1, Jones L. J.1,2, Kasperaviciute D.1,2, Kayikci M.1, Lahnstein L.1, Lawson K.1, Leigh S. E. A.1, Leong I. U. S.1, Lopez F. J.1, Maleady-Crowe F.1, Mason J.1, McDonagh E. M.1,2, Moutsianas L.1,2, Mueller M.1,2, Murugaesu N.1, Need A. C.1,2, Odhams C. A.1, Patch C.1,2, Perez-Gil D.1, Polychronopoulos D.1, Pullinger J.1, Rahim T.1, Rendon A.1, Riesgo-Ferreiro P.1, Rogers T.1, Ryten M.1, Savage K.1, Sawant K.1, Scott R. H.1, Siddiq A.1, Sieghart A.1, Smedley D.1,2, Smith K. R.1,2, Sosinsky A.1,2, Spooner W.1, Stevens H. E.1, Stuckey A.1, Sultana R.1, Thomas E. R. A.1,2, Thompson S. R.1, Tregidgo C.1, Tucci A.1,2, Walsh E.1, Watter S. A.1, Welland M. J.1, Williams E.1, Witkowska K.1,2, Wood S. M.1,2, Zarowiecki M.1.
1Genomics England, London, UK
2William Harvey Research Institute, Queen Mary University of London, London, EC1M 6BQ, UK.
Author Contributions
ZC, WYY, and HH designed the study. ZC, WYY, SE, RS, FB, AF, and TB performed experimental analyses for the study. ZJ provided pathological interpretation and analysis of samples from QSBB. ZC, AT, PS, SAGT, KIG, DZ, JV, and MR carried out either the haplotype analyses, analyses of Genomics England data, provided by GERC and other data analyses. JH, TR, TL, MD, DWD, KAJ, EG, GGK, GH, DBR, IB PT, ASW, NCF, NWW, AJL, and MJH all provided pathological samples, or patient data. HH, ZC, WYY, and JV conceived and designed the study. HH, JV, and MR supervised the project. All authors discussed the results and contributed to the final manuscript.
Conflict of Interest
The authors declare no competing interests.




