Hand-onset weakness is a common feature of ALS patients with a NEK1 loss-of-function variant
Funding Information
This work was supported by the Ministry of Science and Technology, Taiwan (107-2314-B-075-014-MY3), Taipei Veterans General Hospital (V109C-060) and Taiwan Motor Neuron Disease Association.
Abstract
Objective
The NEK1 gene has been recently implicated in amyotrophic lateral sclerosis (ALS). This study aims to assess the influence of NEK1 variants on the occurrence of ALS and investigate the spectrum and clinical features of NEK1 loss-of-function (LOF) variants in a Taiwanese ALS cohort.
Methods
We screened 325 unrelated ALS patients for coding variants in NEK1 by targeted resequencing and queried the Taiwan Biobank database for NEK1 coding variants in 1000 Taiwanese healthy individuals. The clinical features of the patients with a NEK1 LOF variant were analyzed.
Results
Six patients and two healthy individuals carried NEK1 LOF variants. The rare missense variants with minor allele frequencies <0.1% in Taiwanese population were present in 2.8% of the ALS patients and 1.6% of the healthy subjects. NEK1 LOF variants, but not rare missense variants, are significantly enriched in the ALS patients (P = 0.0037 and 0.24, Fisher’s exact test). The odds ratio of an individual carrying a NEK1 LOF variant to develop ALS is 9.39 (95% confidence interval: 1.88–46.7). All the six patients carrying a NEK1 LOF variant had a hand-onset ALS with an onset age from 52 to 64 years. Comparing with ALS patients without a NEK1 LOF variant, patients with a NEK1 LOF variant tend to have a hand-onset disease (P = 0.0008, Fisher’s exact test).
Interpretation
Our study supports the pathogenic role of NEK1 LOF variants and demonstrates their spectrum and clinical features in a Taiwanese cohort with ALS.
Introduction
Amyotrophic lateral sclerosis (ALS) is a devastating neurodegenerative disorder affecting both upper and lower motor neurons, resulting in constantly progressive weakness and muscle atrophy, bulbar function impairment and consequently respiratory failure with a short survival of 3–5 years.1 Approximately 10% of ALS patients inherit their diseases form their parents, indicating that genetic factors substantially contribute to ALS pathogenesis. To date, more than 30 genes have been implicated in ALS pathogenesis. But only a few of them, including C9ORF72, SOD1, FUS, TARDBP, VCP, and TBK1, have been clearly demonstrated to account for a significant number of patients with ALS.2-4 The roles of several newly identified ALS disease genes, such as NEK1,5-8 are yet to be fully understood because the relevant studies are relatively sparse.
The NEK1 gene has been recently implicated in amyotrophic lateral sclerosis (ALS). Loss-of-function (LOF) variants in NEK1 were firstly found strongly associated with ALS in a large-scale whole-exome sequencing study. Cirulli et al. analyzed and compared the whole-exome data of 2843 ALS patients with those of 4310 controls, and additionally verified their findings using a replication cohort consisting of 1318 ALS cases and 2371 controls.5 They found that dominant NEK1 LOF variants were present in 0.835% of cases and 0.091% of controls, suggesting NEK1 as a novel ALS gene with a P-value of 3.2 × 10−9.5 The strong association between NEK1 and ALS was further confirmed in another study by Kenna et al.6 They performed whole-exome analyses in 1022 index cases of familial ALS (FALS) and 7315 controls, and identified a significant association between NEK1 LOF variants and FALS risk. Moreover, they found the NEK1 p.Arg261His variant also as a risk factor of ALS.6 NEK1 encodes the never in mitosis A (NIMA)-related kinase 1 (Nek1), which is a member of Serine/threonine-protein kinase Nek family. The functions of Nek1 include cell cycle control, DNA double-strand break repair, ciliogenesis and mitochondrial membrane permeability regulation.9-11 In the modulation of the DNA damage repair pathway, Nek1 would interact with the chromosome 21 open reading frame 2 (C21orf2) protein,12 which is encoded by the ALS risk gene C21ORF2.13 Interestingly, proteomic analysis showed that Nek1 was associated with ALS2 and VAPB proteins,5 the corresponding genes of which are both well-known ALS disease genes.2, 3
To further understand the role of NEK1 in ALS, we screened 325 unrelated ALS patients for coding variants in NEK1 by targeted resequencing and queried the Taiwan Biobank database for NEK1 coding variants in the 1000 Taiwanese healthy individuals. We analyzed the influence of NEK1 variants on the occurrence of ALS. In addition, we also demonstrated the clinical features, the frequency and spectrum of NEK1 LOF variants in a Taiwanese cohort with ALS.
Methods
Patients
Three hundred and twenty-five unrelated individuals with ALS diagnosed by the revised EL Escorial criteria were enrolled into this study.14 All participants were of Han-Chinese descent and were recruited from the Neurology Service of Taipei Veterans General Hospital, Taiwan. Among the 325 ALS patients, 17 have a SOD1 mutation, 15 have a C9ORF72 GGGGCC hexanucleotide expansion, 12 have a TARDBP mutation, seven have a FUS mutation, two have a CCNF mutation, two have an ATXN2 intermediate-length CAG repeat expansion (32 and 33 repeats) and another three carry a single mutation in OPTN, MATR3, or TBK1 each (Table S1). The remaining 267 patients were not found to have any pathogenic mutation after screening 17 ALS causal genes, including SOD1, C9ORF72, TARDBP, FUS, ATXN2, OPTN, VCP, UBQLN2, SQSTM1, PFN1, HNRNPA1, HNRNPA2B1, MATR3, CHCHD10, TUBA4A, TBK1, and CCNF.15-18 The mean age at disease onset of the ALS cohort was 54.3 years (range 19–89). Thirty-nine patients (12%) had a family history of ALS. Seventy patients (21.5%) were affected by a bulbar-onset ALS and 140 patients (43.1%) had an upper limb-onset disease. Hand-onset ALS was reported in 101 patients (31.1%) and was defined as ALS with the first sign appearing in the hand, usually presenting as difficulty with simple tasks such as buttoning a shirt, using chopsticks, or writing. Peripheral blood samples were collected from the patients after obtaining written informed consents. The protocols for this study were approved by the Institutional Review Board of Taipei Veterans General Hospital.
Genetic analyses
Genomic DNA was extracted from peripheral blood leukocytes using a QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). Mutational analysis of NEK1 was performed by utilizing a targeted resequencing panel covering the whole-coding regions of NEK1. The samples were sequenced on an Illumina MiSeq platform (Illumina, San Diego, CA). All sequenced reads were mapped to the Human Genome version 19 (hg19/GRCh37). The BaseSpace pipeline (https://basespace.illumina.com/) and the IlluminaVariantStudio software (http://variantstudio.software.illumina.com/) were utilized to do variant calling and annotate variants, respectively. The reference coding sequence of NEK1, NM_001199397.3, was used for annotating the variants.
For the NEK1 coding variants identified in the ALS patients, we first checked their frequencies in general populations using the Taiwan Biobank database (TaiwanView: taiwanview.twbiobank.org.tw/search) and the genome Aggregation Database (gnomAD, version r2.0.2; http://gnomad.broadinstitute.org). Taiwan Biobank is a nationwide research database formed to collect blood samples and biomedical information of 200,000 Taiwanese participants using a population-based design.19 Taiwan Biobank has released whole-genome sequence data generated by Illumina Hiseq platforms from 1000 unrelated Taiwanese healthy people. The presence of LOF variants and rare missense variants were further confirmed by Sanger sequencing on the patients’ genomic DNA. The LOF variants were defined as nonsense, frameshift, initiation codon lost, and splice donor/acceptor sites variants, and the rare missense variants were designated for those with a minor allele frequency (MAF) lesser than 0.1% in the 1000 Taiwanese control genomes. Then, we calculated the overall frequencies of all the NEK1 LOF variants and rare missense variants identified in the 1000 healthy Taiwanese genomes from the Taiwan Biobank database as well as those identified in the ALS patients. Fisher’s exact tests were utilized to compare the distributions of NEK1 LOF variants or rare missense variants between the ALS patients and the healthy control genomes, respectively. The frequencies of hand-onset diseases were also compared between the ALS patients with and without a NEK1 LOF mutation. The odds ratio (OR) and 95% confidence interval (CI) was calculated to estimate the risk for an individual carrying a NEK1 LOF mutation or a rare missense mutation to develop ALS.
Results
Genetic analyses
Mutational analyses of NEK1 in the 325 ALS patients revealed six LOF variants in six (1.8%) individuals and eight rare missense variants in nine (2.8%) patients (Table 1).
| cDNA1 | Predicted protein | dbSNP153 | Allele frequency in gnomAD | ALS, n = 325 | Controls, n = 1000 | Fisher’s exact test | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Loss-of-function variants | ||||||||||
| Patient-only variants | ||||||||||
| c.1127_1128delAA | p.Lys376ThrfsTer7 | — | — | 1 | — | |||||
| c.1491delT | p.Pro498LeufsTer10 | — | — | 1 | — | |||||
| c.1897dupA | p.Ile633AsnfsTer28 | — | 0 | 1 | — | |||||
| c.1992delA2 | p.Val665CysfsTer342 | rs775849720 | 2.94E-05 | 1 | — | |||||
| c.2640delA | p.Val881TyrfsTer8 | rs760894879 | 1.05E-05 | 1 | — | |||||
| Variants present in both patients and controls | ||||||||||
| c.2588-2A > G | — | rs201769828 | 1.19E-04 | 1 | 1 | |||||
| Controls-only variants | ||||||||||
| c.1977T > A | p.Tyr659Ter | — | — | — | 1 | |||||
| Total | 6 (1.8%) | 2 (0.2%) | P = 0.0037 | |||||||
| Rare missense variants | ||||||||||
| Patient-only variants | ||||||||||
| c.686A > G | p.Tyr229Cys | rs61737748 | 3.46E-04 | 1 | — | |||||
| c.773T > C | p.Ile258Thr | — | — | 1 | — | |||||
| c.773T > G | p.Ile258Arg | — | — | 1 | — | |||||
| c.1586A > G | p.Glu529Gly | rs781633610 | 8.20E-06 | 1 | — | |||||
| c.1822C > T | p.Arg608Cys | rs749461218 | 1.38E-05 | 2 | — | |||||
| c.1998G > C | p.Trp666Cys | rs746590014 | 8.18E-06 | 1 | — | |||||
| c.2955A > C | p.Gln985His | — | — | 1 | — | |||||
| Variants present in both patients and controls | ||||||||||
| c.460A > T | p.Asn154Tyr | rs756418625 | 1.34E-05 | 1 | 1 | |||||
| Controls-only variants | ||||||||||
| c.770T > G | p.Phe257Cys | — | — | — | 1 | |||||
| c.1625G > A | p.Arg542Gln | rs1032792680 | 1.61E-05 | — | 1 | |||||
| c.1634T > C | p.Met545Thr | rs760674667 | 2.1E-05 | — | 1 | |||||
| c.1703G > A | p.Arg568Lys | rs1561319838 | — | — | 1 | |||||
| c.1769G > A | p.Arg590Lys | — | — | — | 1 | |||||
| c.1823G > A | p.Arg608His | rs551275723 | 6.46E-05 | — | 1 | |||||
| c.1978G > A | p.Glu660Lys | rs1458058029 | — | — | 1 | |||||
| c.1982G > A | p.Arg661Lys | — | — | — | 1 | |||||
| c.1984G > A | p.Glu662Lys | — | — | — | 1 | |||||
| c.2183A > C | p.Gln728Pro | — | — | — | 1 | |||||
| c.2195A > G | p.Asn732Ser | rs755259769 | 1.24E-05 | — | 1 | |||||
| c.2287A > C | p.Thr763Pro | — | — | — | 1 | |||||
| c.2736G > A | p.Met912Ile | rs750788096 | 1.62E-05 | — | 1 | |||||
| c.3149C > T | p.Pro1050Leu | — | — | — | 1 | |||||
| c.3289G > A | p.Val1097Ile | rs374890006 | 8.11E-05 | — | 1 | |||||
| Total | 9 (2.8%) | 16 (1.6%) | P = 0.24 | |||||||
- ALS, amyotrophic lateral sclerosis; dbSNP153: Single Nucleotide Polymorphism database, build 153; gnomAD, the Genome Aggregation Database; n, numbers of variant carriers in the ALS cohort or 1000 controls from Taiwan Biobank (https://www.twbiobank.org.tw).
- 1 The reference coding sequence of NEK1, NM_001199397.3, was used for annotating the variants.
- 2 In a homozygous form; other variants in the table are heterozygous.
The six NEK1 LOF variants include c.1127_1128delAA (p.Lys376Thrfs*7), c.1491delT (p.Pro498Leufs*10), c.1897dupA (p.Ile633Asnfs*28), c.1992delA (p.Val665Cysfs*34), c.2640delA (p.Val881Tyrfs*8), and c.2588-2A > G (Fig. 1). Among these LOF variants, NEK1 p.Val665Cysfs*34 was present in a homozygous form in the patient and all the others are heterozygous. Two patients carrying the same NEK1 rare missense variant, p.Arg608Cys, also had a heterozygous TARDBP p.Met337Val mutation. All the other thirteen patients with NEK1 variants did not have any known mutation in other ALS disease genes.
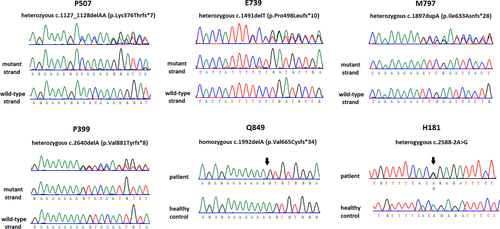
From the genomic data of 1000 Taiwanese healthy individuals in the Taiwan biobank, we found two LOF variants in two (0.2%) individuals and 16 rare missense variants in 16 (1.6%) individuals (Table 1). Comparing the frequencies of NEK1 LOF variants or rare missense variants in the ALS patients with those in the 1000 Taiwanese individuals, NEK1 LOF variants but not rare missense variants are significantly enriched in the ALS patients (P = 0.0037 and 0.24, respectively). The odds ratio for an individual carrying a NEK1 LOF variant to develop ALS is 9.39 (95% confidence interval: 1.88–46.7). In contrast, harboring a NEK1 rare missense variant did not significantly increase the risk to develop ALS (odds ratio: 1.75, 95% confidence interval: 0.77–4).
Clinical information of the patients carrying NEK1 LOF variants
The clinical and genetic features of the six ALS patients harboring NEK1 LOF variants are summarized in Table 2. These patients neither suffered from dementia nor had a family history of ALS. Patient P507 was heterozygous for the NEK1 p.Lys376Thrfs*7 mutation. Her disease started with left hand weakness at age 63 years. The symptoms deteriorated rapidly, and she developed four limbs weakness within 1 year. Neurological examinations at age 65 years revealed weakness and atrophy with fasciculation in bilateral upper extremities (muscle strength of 4- to 4+/5 according to the Medical Research Council-scale), a milder degree of involvement in the bilateral lower limbs (4 to 4+/5), diminished deep tendon reflexes (DTRs), and normal cognitive function. The motor symptoms were more severe on left side limbs and distal parts. Her scores on the Amyotrophic Lateral Sclerosis Functional Rating Scale-revised (ALSFRSr) were 33 at age 65 years and 22 at age 66 years. She died of respiratory failure at age 67 years.
| Patient | Sex | Mutation | Hetero- or homozygous | Age of onset, y | Site of onset | Age at first evaluation, y | Survival, m | Clinical phenotype | Dementia |
|---|---|---|---|---|---|---|---|---|---|
| P507 | F | p.Lys376Thrfs*7 | Heterozygous | 63 | left hand | 65 | 46 | LMN | No |
| E739 | M | p.Pro498Leufs*10 | Heterozygous | 56 | right hand | 58 | 98 | UMN/LMN | No |
| M797 | M | p.Ile633Asnfs*28 | Heterozygous | 54 | left hand | 55 | 13 | UMN/LMN | No |
| P399 | M | p.Val881Tyrfs*8 | Heterozygous | 64 | right hand | 66 | 43 | UMN/LMN | No |
| Q849 | M | p.Val665Cys*34 | Homozygous | 55 | left hand | 55 | >181 | LMN | No |
| H181 | M | c.2588-2A > G | Heterozygous | 52 | right thumb | 55 | >120 | LMN | No |
- ALS, amyotrophic lateral sclerosis; y, years; m, months; UMN, upper motor neuron signs; LMN, lower motor neuron signs.
- 1 > 18 means that more than 18 months have passed since the symptom onset and the patient was still alive on the last follow-up.
Patient E739 had a heterozygous NEK1 p.Pro498Leurfs*10 mutation with an onset symptom of right hand weakness at age 56 years. The symptoms progressed to bilateral upper limbs, bulbar region, and then lower limbs sequentially within 2 years. He received tracheostomy at age 59 years and died of an out-of-hospital cardiac arrest at age 64 years.
Patient M797 was heterozygous for the NEK1 p.Ile633Asnfs*28 mutation. He had a disease onset at age 54 years with left hand weakness. The symptoms rapidly extended to bilateral upper limbs, bulbar region, and then lower limbs within 6 months. Neurological examinations at age 55 years revealed severe weakness and atrophy in bilateral upper extremities (2 to 3/5), a milder degree of involvement in the bilateral lower limbs (4 to 4+/5), tongue atrophy with fasciculation, dysarthria, brisk DTRs in the upper limbs and normal knee and ankle DTRs. He died of respiratory failure at age 55 years.
Patient P399 had a heterozygous NEK1 p.Val881Tyrfs*8 mutation and the disease began with weakness in the right hand and fingers at age 64 years. The weakness progressed to proximal right upper limb first and then right lower limb within 2 years. Neurological examinations at age 66 years revealed Mills syndrome, presenting with severe weakness in the right upper limb (0 to 1/5) and moderate weakness in the right lower limb (4- to 4/5), atrophy over right hand, increased deep tendon reflexes, right extensor plantar response, and normal cognitive function. Cervical MRI at age 66 showed mild cervical spondylosis, which mainly affected C5-6 level without cord or root compression. His ALSFRSr score was 33. The motor symptoms spread to left side limbs presenting with mild weakness (4+/5) within 4 months after first evaluation. Then, he was lost to follow-up for a period and died of pneumonia at age 68 years.
Patient Q849 is homozygous for the NEK1 p.Val665Cys*34 mutation. He had a disease onset at the age of 55 years with left hand weakness. Neurological examinations at 6 months after the disease onset revealed weakness and atrophy in bilateral upper limbs, which was more severe in distal parts of extremities (4- to 4+/5), and decreased deep tendon reflexes. He had dyspnea on exertion at age 56 years and received tracheostomy with ventilator support soon at the 17th month after the disease onset. His ALSFRSr scores were 33 at first evaluation and downhill to 2 at one year later. He denied that his parents had a consanguineous marriage.
Patient H181 had a heterozygous NEK1 c.2588-2A > G mutation with an onset symptom of weakness in the right thumb at age 52 years. The symptoms progressed to bilateral upper limbs and then bulbar region and lower limbs within 3 years. He received percutaneous endoscopic gastrostomy feeding and tracheostomy ventilation at age 55 years. Neurological examinations at age 62 years revealed total paralysis and atrophy of four limbs (0 to 1/5), severe tongue atrophy with frequent fasciculation, and decreased deep tendon reflexes.
All the six patients carrying NEK1 LOF mutations had a hand-onset ALS. However, only 101 of the 325 ALS patients in the study had hand weakness as the initial clinical presentation. Comparing with ALS patients without NEK1 LOF mutations, ALS patients with a NEK1 LOF mutation tend to have a hand-onset disease (100% vs. 29.8%, P = 0.0008). The clinical and genetic information of the ALS patients harboring NEK1 rare missense variants are summarized in Table S2.
Discussion
In this study, we evaluated the effects of NEK1 mutations on ALS by comparing the frequencies of NEK1 LOF variants and rare missense variants in Taiwanese ALS patients with those in healthy individuals. We also characterized the clinical and genetic features of the Taiwanese ALS patients carrying NEK1 LOF variants. There are four major findings from this study. First, NEK1 LOF variants are significantly enriched in the Taiwanese ALS patients. Second, harboring a NEK1 rare missense variant does not significantly increase the risk to develop ALS. Third, approximately two percent of ALS cases in Taiwan may be attributed to NEK1 LOF variants. Finally, comparing with ALS patients without NEK1 LOF variants, patients with NEK1 LOF variants tend to have a hand-onset disease. These findings may have the following implications.
First of all, our study demonstrates a clear association between NEK1 LOF variants and ALS. In the studied population, Taiwanese individuals with a NEK1 LOF variant pose a 9.34-fold risk to develop ALS, whereas harboring a NEK1 rare missense variant does not significantly increase the risk of having ALS. Similar findings were also demonstrated in Caucasian populations recently by several large-scale whole-exome sequencing studies, in which NEK1 LOF were significantly associated with ALS risks.5-8 The association between ALS and NEK1 LOF mutations in multiple populations suggests that NEK1 LOF variants are pathogenic and NEK1 haploinsufficiency may contribute to ALS pathogenesis. It is still unclear how NEK1 haploinsufficiency links to ALS pathogenic. Further studies are warranted to elucidate the relevant molecular mechanism.
Second, NEK1 LOF variants may account for a certain number of ALS patients. This study identified NEK1 LOF variants in six of 325 ALS patients, implying that the prevalence of NEK1 LOF variants in Taiwanese ALS patients is 1.8%. Several recent NEK1 studies revealed that the prevalence of NEK1 pathogenic variants in sporadic and familial ALS patients ranged from 0.8% to nearly 3% in different populations.5-7, 20, 21 Comparing to the prevalence of other disease genes in ALS populations, the frequency of LOF variants in NEK1 is not rare. The percentage of Taiwanese ALS patients carrying a NEK1 LOF variant is lower than the rates of ALS patients harboring a mutation in SOD1, C9ORF72, TARDBP, and FUS, but higher than those of individuals with mutations in other ALS disease genes.
Third, NEK1 LOF variants may be pathogenic variants with reduced penetrance for ALS. In this study, lack of family history of the six ALS patients carrying the NEK1 LOF variants suggests an incomplete penetrance of these variants. Reduced penetrance of NEK1 LOF variant has also been demonstrated in another study reporting an unaffected sibling of ALS patients carrying the NEK1 p.Ser1036* variant.7 Furthermore, the frequencies of healthy controls carrying the NEK1 LOF variants are 0.2% in the Taiwanese population and 0.06–0.16% in the Caucasian populations.5-7, 20 The relatively high prevalence of NEK1 LOF variants in control subjects implies that the penetrance of NEK1 LOF variants is not high. Since ALS is a devastating disease, it is still important to elucidate the NEK1-related pathogenic mechanism and develop tailored strategies to prevent and treat ALS in individuals carrying a NEK1 pathogenic variant.
Fourth, hand-onset weakness seems to be a common feature of ALS patients harboring a NEK1 LOF variant. Brenner et al. reported four familial ALS patients with NEK1 LOF variants from three families.7 Among them, two had a hand-onset disease and another one patient had upper and lower motor neuron signs beginning from left arm.7 Another study from Beijing China reported three ALS patients with NEK1 LOF variants and showed that the two patients with available clinical information both had a hand-onset disease.21 In our ALS cohort, all the six ALS patients carrying NEK1 LOF variants manifested a hand-onset disease, whereas lesser than one third of the remaining 319 ALS patients without a NEK1 LOF variant had symptoms beginning from hands. It remains unclear why NEK1 LOF variants are associated with hand-onset ALS. Low cervical anterior horn cells might have a higher demand on normal Nek1 functioning than lower motor neurons in other regions. Phenotypic preferences have also been observed in ALS associated with mutations in other disease genes, such as SOD1 mutations correlating with flail leg phenotype and C9ORF72 expansions associating with bulbar phenotype.22
In conclusion, we identifies six ALS patients with NEK1 LOF variants in 325 unrelated Taiwanese ALS patients. All the patients had a hand-onset ALS with an onset age ranged from 52 to 64 years. This study characterizes the genetic and phenotypic features of NEK1 LOF variants and underlines their pathogenic role in ALS.
Acknowledgments
We thank the patients who participated in this study. This work was supported by the Ministry of Science and Technology, Taiwan (107-2314-B-075-014-MY3), Taipei Veterans General Hospital (V109C-060) and Taiwan Motor Neuron Disease Association. We also thank the High-throughput Genome Analysis Core Facility of National Core Facility Program for Biotechnology of Taiwan for the genetic analysis service.
Conflict of Interest
The authors have no conflicts of interest to disclose.




