P5CS expression study in a new family with ALDH18A1-associated hereditary spastic paraplegia SPG9
Funding information
Whole-exome sequencing analysis was funded by a “Fondazione del Monte” grant to Professor MS for clinical exome sequencing applied to the diagnosis of ultrarare/orphan inherited diseases. The Genetic Bank (Biobanca del Laboratorio di Genetica Umana IRCCS Gaslini), member of the Network Telethon of Genetic Biobanks (Project No. GTB12001), funded by Telethon Italy, provided us with specimens. VR was supported by grants of the Fundación Inocente Inocente and of the Spanish Government (MINECO BFU2017-84264-P).
Abstract
In 2015–2016, we and others reported ALDH18A1 mutations causing dominant (SPG9A) or recessive (SPG9B) spastic paraplegia. In vitro production of the ALDH18A1 product, Δ1-pyrroline-5-carboxylate synthetase (P5CS), appeared necessary for cracking SPG9 disease-causing mechanisms. We now describe a baculovirus–insect cell system that yields mgs of pure human P5CS and that has proven highly valuable with two novel P5CS mutations reported here in new SPG9B patients. We conclude that both mutations are disease-causing, that SPG9B associates with partial P5CS deficiency and that it is clinically more severe than SPG9A, as reflected in onset age, disability, cognitive status, growth, and dysmorphic traits.
Introduction
ALDH18A1 gene-related spastic paraplegias (SPG9A and SPG9B) are extremely rare disorders recognized quite recently.1, 2 They are due to mutations in ALDH18A1 (Fig. 1A), which encodes Δ1-pyrroline-5-carboxylate synthetase (P5CS), an enzyme that catalyzes the initial two steps of ornithine and proline biosynthesis.3, 4 This bifunctional single-chain homo-oligomeric2 protein is composed of glutamate 5-kinase (G5K) and glutamyl-5-phosphate (G5P) reductase (G5PR) components3 (Fig. 1A). Biallelic ALDH18A1 mutations were known to cause a cutis laxa neurocutaneous syndrome5, 6 (ARCL3A, MIM#219150). In 2015–2016, we and others1, 2 reported ALDH18A1 mutations in dominant complicated spastic paraplegia (SPG9A, MIM#601162). A recessive form of SPG9 (SPG9B) associated to biallelic ALDH18A1 mutations was also reported1 (SPG9B, MIM#616586). The effects of ALDH18A1-related disease mutations appear to result from loss of P5CS7-9/decrease of P5CS function1, 2, 5, 6, 10-12, leading to the proposal that dominant mutations cause ALDH18A1 pathologies by a dominant negative effect2, 11, 12. Similarly to Charcot-Marie-Tooth disease due to GDAP1 gene mutations13, it is conceivable that dominant SPG9 mutations could cause a less severe disease phenotype than biallelic recessive ones.
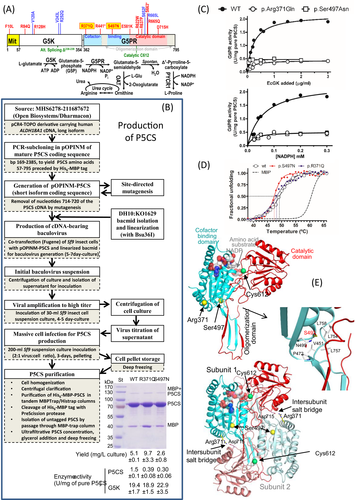
If disease severity in SPG9 has to be correlated with P5CS enzyme activity, it is crucial to have a procedure for directly testing the mutations’ effects on P5CS activity and stability. We describe here this procedure for in vitro production of pure P5CS using an engineered baculovirus–insect cell system, exemplifying the value of this system with two novel ALDH18A1 mutations found in a new family of SPG9B that is also reported here. In addition, we rationalize the observed effects of these mutations using the crystal structure (Protein Database, PDB, entry 2H5G) of the G5PR component of human P5CS. Furthermore, by expanding by one family the meager (just 6 prior families)1, 9, 14, 15 SPG9B database, we conclude on sound bases that SPG9B is generally more severe than SPG9A.
Materials and Methods
See Supplementary Methods for details.
Production of human P5CS
An engineered baculovirus was prepared16-18 (Fig. 1B) by co-transfection of Sf9 cells with bacmid DH10:KO1629 and with a pOPINM plasmid (Clontech) bearing the mature human P5CS coding sequence (Fig. S1). This virus was used for production in Sf9 insect cells of P5CS preceded by a His6-MBP-3C protease cleavable double tag17. Clinical mutations were introduced by site-directed mutagenesis of the pOPINM derivative carrying the P5CS cDNA. Doubly-tagged P5CS purified by tandem affinity chromatography with MBP- and His-trap columns (GE Healthcare) was freed from both tags by PreScission protease (GE Healthcare) digestion followed by nontagged P5CS isolation by a final affinity chromatography step.
Enzyme activity and stability assays
Spectrophotometric activity assays at 25°C were used2, monitoring at 340 nm the oxidation of either NADPH in the complete P5CS reaction or the G5PR partial reaction; or the oxidation of NADH in the G5K partial reaction, coupling ADP production to NADH oxidation using pyruvate kinase and lactate dehydrogenase.
Thermofluor assays19 were used to monitor P5CS protein unfolding with increasing temperature (1°C increase/min). Here, Tm is the temperature giving a fluorescence increase of 50% of the maximum increase attained.
Clinical and genetic studies
Consent was obtained for DNA sequencing, for using the results in research, and for this publication. Retrospective clinical and laboratory information gathered for patient care was used, precluding the need for ethics committee approval. We complied with the ethical principles of the Helsinki Declaration.
Biallelic ALDH18A1 mutations were identified in the index patient by whole-exome sequencing (WES). Sanger sequencing (Fig. S2) was used for confirmation and for familial segregation analysis.
Results and Discussion
See also Supplementary Results.
Production of recombinant human P5CS
We recombinantly produced human P5CS as schematized in Figure 1B in an engineered baculovirus–insect cell system. The doubly-tagged P5CS produced could be purified easily by tandem affinity chromatography followed by tag removal (PreScission protease) and a second round of affinity purification that frees P5CS from residual non-cleaved tagged P5CS and from the excised tags. Using this approach, mg amounts of homogeneous largely pure (Fig. 1B, bottom image and yield below) P5CS were obtained from just 200 mL of insect cell culture. The MBP tag was crucial for P5CS solubility, although tag removal was essential for eliciting P5CS activity (data not shown).
Production of mutant forms of P5CS
The p.Arg371Gln and the p.Ser497Asn novel P5CS mutations (reported here in a new SPG9B family, see below) caused no gross P5CS misfolding, since both mutant forms were produced and purified with similar yield as wild-type P5CS (Fig. 1B, bottom image and yield below). Similar success was obtained in a prior study2 with the pilot application of this method (without describing it) to the p.Val243Leu and p.Arg252Gln mutations found in SPG9A families. Thus, in principle, it appears that this procedure has general value for generating any clinical P5CS missense mutation.
The novel SPG9B mutations p.Arg371Gln and p.Ser497Asn decrease but do not abolish P5CS activity
p.Arg371Gln and p.Ser497Asn did not abolish P5CS activity although both of them caused 75–80% decreases in this activity (Fig. 1B, P5CS activity below gel image). These decreases in activity account for disease causation. They also show that the SPG9B phenotype is associated with a substantial level of residual P5CS activity. In contrast, some patients with the ALDH18A1-caused cutis laxa phenotype carried biallelic null mutations and exhibited total absence of P5CS protein.7, 8
In agreement with the mapping of the mutations in the G5PR component of P5CS, these mutations did not substantially affect the G5K partial activity (Fig. 1B, G5K activity below gel image). In fact, they selectively decreased approximately fivefold the G5PR partial activity measured at saturation of any of the two substrates of this reaction, G5P and NADPH (Fig. 1C, values at high concentrations of these substrates; G5P levels are proportional to the amount of EcG5K added, see Supplementary Methods). As Km values for these two substrates were not increased (Fig. 1C), the decrease in the activity was purely a Vmax effect.
None of these two mutations affected importantly P5CS thermal stability, since although they decreased Tm values, the decrease was quite small (1–2°C, Fig. 1D).
Structure-based rationalization of the mutations’ effects
The failure of the p.Arg371Gln and p.Ser497Asn mutations to increase the Km values for G5P and NADPH in the G5PR reaction agrees with the mapping of Arg371 or Ser497 in the structure of the G5PR component of P5CS, away from the inferred binding sites for these substrates (Fig. 1E, upper panel). The magnitude of the decrease in Vmax due to the mutations militates against a catalytic role of Arg371 and Ser497, two residues that sit far from the catalytic Cys612 (Fig. 1E, upper panel). It fits better the triggering by the mutations of more indirect effects, possibly due to changes in the equilibrium and/or the kinetics for the opening and closing of the two globular domains of the G5PR component inferred to take place during catalysis.20 By making a salt bridge with Asp715 of the other subunit of the G5PR dimer (Fig. 1E, bottom panel), Arg371 stabilizes the open conformation. The Arg371Gln substitution abolishes such salt bridge and prevents this stabilization. Ser497 sits at the junction of the cofactor-binding domain, the catalytic domain and the oligomerization domain of the G5PR component (Fig. 1E, upper panel). This serine makes hydrophobic interactions in the interdomain regions that involve the beginning of the protruding oligomerization domain (detailed in the middle panel of Fig. 1E), holding together these elements and anchoring them on the cofactor-binding domain. Mutation to Asn could disturb this anchoring, perhaps altering the dynamics rather than the equilibrium between open and closed forms.
A novel family presenting the recessive form of ALDH18A1-related spastic paraplegia
We diagnosed SPG9B in a 37-year-old male (see details in Supplementary Results) referred to us for a history of slowly progressive gait and motor disability due to spastic paraplegia, which led him over many years to the use of a walker and, from his mid-thirties, of a wheelchair. The patient showed intellectual disability, microcephaly, short stature, dysmorphic features (predominating facial ones), and irritative electroencephalographic findings. ALDH18A1 missense mutations in compound heterozygosity were identified by WES and were confirmed by Sanger sequencing (Fig. S2). A brother of the patient showing a similar phenotype with onset at 3 years of age, in addition presenting seizures, was proven by Sanger sequencing to carry the same mutations as his brother. Each parent was confirmed for one variant, while a sister did not carry either (Fig. 2A). Family history was negative for intellectual or motor disabilities.
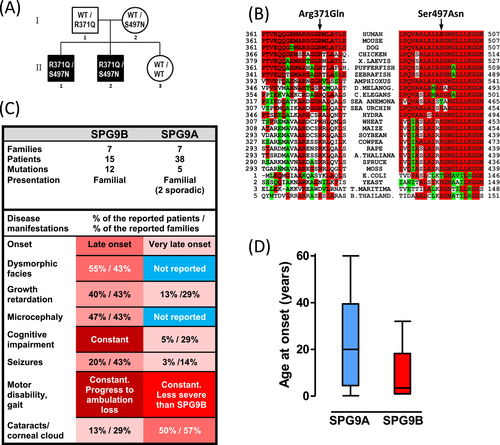
The ALDH18A1 variants identified in the patients (Table 1) were c.1112G>A and c.1490G>A. The first, c.1112G>A, p.(Arg371Gln), affects a CpG island (hypermutable islands) and was found in heterozygous state in 7 out of 282,858 alleles in the GnomAD database. The second, c.1490G>A, p.(Ser497Asn), was unreported in databases. Both mutations were absent from our local database of WES data for nearly 1000 individuals. Neither was predicted to affect splicing (Human Splicing Finder server, http://www.umd.be/HSF3). Pathogenicity of the variants was supported by low or no presence of these variants in genomic variation databases, conservation of the bases (PhyloP scores, Table 1) and amino acids at these positions (Fig. 2B), and by unanimous predictions of disease causation by PolyPhen2, MutPred2, and MutationTaster 2 (Table 1), and, finally, by the above-reported experimental assays with the recombinantly produced protein.
| Nucleotide changea | Protein changeb | Amino acid in P5CS or in isolated microbial G5PRc | Domain of G5PR component | Pathogenicity server prediction | Base conservation | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Polyphen 2d | MutPred2e | Mutation taster 2f | PhyloPg | |||||||
| Animals | Plants | Microbial | Prediction | Score | Score | Score | ||||
| c.1112G> A | p.Arg371Gln | R | R | R/k | cofactor binding | Probably damaging | 1 | 0.623 | Disease-causing | 5.035 |
| c.1490G> A | p.Ser497Asn | S/t | S | S/t | cofactor binding | Probably damaging | 1 | 0.791 | Disease-causing | 2.746 |
- a GeneBank (https://www.ncbi.nlm.nih.gov/nucleotide/) reference sequences for human ALDH18A1 gene, its mRNA (isoform 1, long form) and protein (long form), NG_012258.1, NM_002860.3 and NP_002851.2, respectively. Nucleotide numbering uses + 1 as the A of the ATG translation initiation codon (codon1).
- b Uniprot KB (https://www.uniprot.org/uniprot/) reference number P54886.
- c Amino acid conservation was determined by sequence alignment using Clustal, (https://www.ebi.ac.uk/Tools/msa/clustalo) of either P5CS from animals or plants or microbial G5PR from 45, 30 or 20 species, respectively. Residues in single letter amino acid code. Non-capitalized letters denote occurrence in low frequency.
- d Polyphen-2 (HumVar-trained dataset; http://genetics.bwh.harvard.edu/pph2) grades the probability of a damaging effect of an amino acid change, as probably damaging, possibly damaging and benign. Highest probability score is 1.
- e The score given by MutPred2 (http://mutpred2.mutdb.org) is the probability that a given amino acid change is deleterious/disease associated.
- f http://www.mutationtaster.org
- g PhyloP measures evolutionary conservation at individual alignment sites providing positive scores at sites that are predicted to be conserved (maximum 6) or negative scores (mínimum −14) when sites are predicted to evolve fast. The PhyloP result was taken from the report made by Mutation Taster
Higher severity of SPG9B than of SPG9A
Our report of a further SPG9B family enlarges the family/patient/mutation database (Figs. 1A and 2C), and provides more evidence and insight into the correlation of the SPG9A and SPG9B phenotypes with their associated genotypes (Fig. 2C). These comparisons (Fig. 2C and D) strongly suggest that SPG9A, caused by dominant mutations, exerts less severe effects that the combined effects of the two recessive mutations found in SPG9B patients, who generally exhibit an earlier onset (Fig. 2D) and a progression to higher disability than SPG9A patients (Fig. 2C). They invariably present cognitive impairment and, frequently, short stature, microcephaly, seizures, and dysmorphic features. These features have been reported less frequently or not at all in SPG9A patients (Fig. 2C). Only early cataracts seems to be more frequent in SPG9A than in SPG9B (Fig. 2C); however, more case reports would be needed to draw conclusions regarding cataract frequency.
Conclusions
The in vitro P5CS production system reported here is a highly useful tool to clarify the effects of individual P5CS mutations and thus to establish the molecular mechanisms of disease causation in ALDH18A1-related disorders. The use of this system has shown that the novel P5CS mutations p.Arg371Gln and p.Ser497Asn reported here in a new SPG9B family are disease causing, importantly decreasing P5CS enzyme activity. However, since they do not completely inactivate this enzyme, SPG9B is associated to partial rather than total P5CS deficiency. With the increased SPG9B family/patient database after addition of this new family, the dominant and recessive form of SPG9 can be compared on sound bases, leading to the conclusion that SPG9B is more severe than SPG9A, having a lower onset age, attaining greater disability, presenting constant impairment of cognition and more frequent growth restriction and dysmorphic traits.
Acknowledgments
We thank Nick S Berrow (IRB-Parc Cientific, Barcelona) and Prof. Ian M Jones (University of Reading, UK) for the DH10:KO1629 bacmid, and patients and family members who participated in this study, as well as the American Spastic Paraplegia Foundation for the EP grant ”Understanding Hereditary Spastic Paraplegia: in vivo models to identify pathogenetic mechanism and therapeutic targets for SPG9,” which was partially used for the research in this work. Whole-exome sequencing analysis was funded by a “Fondazione del Monte” grant to Professor MS for clinical exome sequencing applied to the diagnosis of ultrarare/orphan inherited diseases. The Genetic Bank (Biobanca del Laboratorio di Genetica Umana IRCCS Gaslini), member of the Network Telethon of Genetic Biobanks (Project No. GTB12001), funded by Telethon Italy, provided us with specimens. VR was supported by grants of the Fundación Inocente Inocente and of the Spanish Government (MINECO BFU2017-84264-P).
Conflict of Interest
The authors declare that they do not have any commercial or financial conflict of interests.
Authors Contributions
MS, EP, and VR conceived and designed the study; PM contributed with the Exome Sequencing and data analysis; CM-M, JME-H, and VR produced and functionally characterized P5CS, either wild-type or mutant, and carried out the structural analysis; FF, FF, DM, and CDV contributed to the patients clinical description and analysis of results. EP, MS, VR, and CM-M wrote the manuscript, and all the authors read it and made contributions to improve its writing.




