A Prokaryotic (6-4) Photolyase with a DMRL Chromophore and an Iron–Sulfur Cluster
Abstract
Photolyases are flavoproteins that repair UV-damaged DNA using the energy of sunlight. The flavin adenine dinucleotide (FAD) chromophores of photolyases and the related cryptochromes are bound to the chromophore pocket in a U-shaped conformation. Often, a second cofactor serves as antenna chromophore in photolyases. This can be methenyltetrahydrofolate (MTHF), 8-hydroxy-7,8-didemethyl-5-deazariboflavin (8-HDF), FAD, or flavin mononucleotide (FMN). Two kinds of DNA lesions are produced by UV light, cyclobutan-pyrimidine dimers (CPDs) or (6-4) photoproducts. These lesions are repaired by two different types of photolyases, namely the CPDs and (6-4) photolyases.
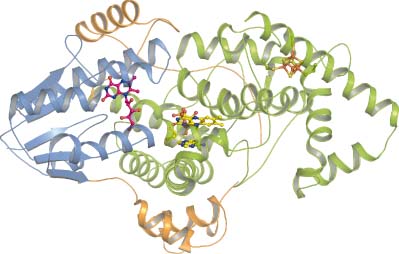
Recently, a new phylogenetic group of the photolyase cryptochrome family termed iron–sulfur bacterial cryptochromes/photolyases (FeS-BCP) was discovered. Two members of this group, CryB from Rhodobacter sphaeroides and ‘photolyase related protein’ (PhrB) from Agrobacterium tumefaciens, carry a new antenna chromophore, 6,7-dimethyl 8- ribityl-lumazin (DMRL), and incorporate an 4Fe-4S cluster. PhrB acts as a (6-4) photolyase with a repair activity that is higher for double-stranded than for single-stranded DNA. There are presently more than 900 prokaryotic PhrB homologs in the database in which the characteristic cysteine residues for incorporation of the Fe-S cluster and other key amino acids are conserved. This new group of (6-4) photolyases with an Fe-S cluster is thus widely distributed among prokaryotes. Unlike in other photolyases, the loop connecting the N-terminal and the C-terminal domains of PhrB interacts with the DNA lesion. A C-terminal extension, which could have a regulatory function, interacts with the DNA. The comparison between PhrB and eukaryotic (6-4) photolyases highlights a conserved His residue which is located next to the DNA lesion. This residue serves probably as proton donor/acceptor during the repair cycle.