IL-4 and IL-13 regulate the induction of indoleamine 2,3-dioxygenase activity and the control of Toxoplasma gondii replication in human fibroblasts activated with IFN-γ
Abstract
The ability of up-regulatory [recombinant (r) IFN-γ, rIFN-β and rTNF-α] and down-regulatory (rIL-4, rIL-10 and rIL-13) cytokines to control the expression of indoleamine 2,3-dioxygenase (INDO) and anti-Toxoplasma activity in the human fibrosarcoma cell line 2C4 was evaluated. Activation of fibroblasts with rIFN-γ, rIFN-β and rTNF-α resulted in augmentation of INDO expression and activity leading to 40.0, 25.0 and 27.0 % inhibition of tachyzoite growth, respectively. An additive effect was observed when host cells were incubated with rIFN-γ plus rTNF-α. With regard to the down-regulatory cytokines we observed that IL-4 as well as IL-13, but not IL-10, induced significant inhibition of IFN-γ-induced control of parasite replication, INDO mRNA expression and tryptophan catabolism. Similarly, IL-4 but not IL-10 inhibited the cell surface expression of HLA-DR and CD2 induced by IFN-γ. Consistent with these findings we were able to detect by reverse transcription-PCR the expression of mRNA for different chains of IL-4 and IL-13 receptors (IL-4Rα, IL-13Rα1 and IL-13Rα2) but not for IL-10 receptor in the 2C4 and other human lung fibroblast cell lines (LL24 and MRC5). Together our results indicate that IL-4 and IL-13, but not IL-10, are implicated in the negative regulation of IFN-γ-induced anti-Toxoplasma activity in human cells from fibroblast lineage.
Abbreviations:
-
- INDO:
-
Indoleamine 2,3-dioxygenase
-
- PPC:
-
Professional phagocytic cells
-
- NPPC:
-
Non-professional phagocytic cells
-
- RT:
-
Reverse transcription
-
- SBE:
-
Signal-binding element of STAT
-
- IRF:
-
Interferon-regulatory factor
-
- TCA:
-
Trichloroacetic acid
-
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
1 Introduction
Several gene products and functions induced by IFN-γ, in either professional phagocytic cells (PPC) or non-professional phagocytic cells (NPPC), have been implicated in resistance to microbial infection. In macrophages, or PPC, the activation by IFN-γ leads to the production of reactive oxygen and nitrogen intermediates 1 – 3, as well as tryptophan degradation 4, 5, all involved in the control of intracellular pathogen replication. The indoleamine 2,3-dioxygenase (INDO) is the enzyme that catalyzes the initial and rate-limiting step of tryptophan catabolism to N-formylkynurenine and kynurenine. The in vitro induction of INDO leads to rapid depletion of intracellular tryptophan and accumulation of its metabolites in the cell supernatant. Restriction of available tryptophan due to degradation by INDO leads to a condition in which cells become starved for tryptophan. Many cell lines are responsive to and produce INDO upon induction by three types of IFN (α, β and γ), although IFN-γ is the most effective 5. Induction of cellular INDO expression has been implicated as a mechanism to account for the anti-proliferative activity of IFN-γ on tumor cells, inhibition of the growth of intracellular pathogens 5 and more recently described as crucial for establishing immune tolerance during pregnancy 6.
In human NPPC the induction of INDO appears to be the main mechanism by which IFN-γ controls the intracellular replication of Toxoplasma gondii tachyzoites. In the absence of tryptophan, an essential amino acid for T. gondii, the pathogen growth becomes restricted. The tryptophan catabolite kynurenine is not toxic for the parasite and the addition of tryptophan to the tissue culture medium restores Toxoplasma growth in these cells 4, 7, 8. The anti-Toxoplasma activity of the INDO induced by IFN-γ has been shown to be potentiated by IFN-γ and TNF-α 9 – 12. Previous studies to define whether IFN-β alone can confer upon human cells the capacity to inhibit or kill T. gondii tachyzoites have yielded conflicting results. In some studies minimal or no effect was observed 13, 14, whereas in others a significant effect was reported 12, 15.
It has been known that IL-4, IL-10, IL-13 and TGF-β are the most effective anti-inflammatory cytokines that antagonize many cellular responses induced by IFN-γ, including different functions displayed by macrophages 2, 16 – 23. However, the role of these regulatory cytokines mediating the negative effect on control of T. gondii growth and INDO expression in NPPC have received scant attention to date. In the present study, we evaluated the ability of up-regulatory (i. e. rIFN-γ, rIFN-β and rTNF-α) and down-regulatory cytokines (i. e. rIL-4, rIL-10 and rIL-13) to control T. gondii replication and tryptophan catabolism in the human fibrosarcoma cell line 2C4. Contrasting with studies performed with PPC 24 – 25, our results favor the hypothesis that IL-4 and IL-13, but not IL-10, are major antagonists of IFN-γ-induced activities, including tryptophan degradation by INDO and microbiostasis in NPPC.
2 Results
2.1 Cytokine effect on intracellular replication of T. gondii
The levels of T. gondii replication in human 2C4 fibroblasts activated with rIFN-γ, rIFN-β and / or rTNF-α are presented in Table 1. The stimulation of 2C4 cells with 100 and 500 IU / ml rIFN-γ resulted in significant inhibition of intracellular tachyzoite replication of 36.4 and 43.8 %, respectively. A smaller but statistically significant inhibitory effect of rTNF-α (60 IU / ml) and rIFN-β (1000 IU / ml) on T. gondii growth was observed in different experiments. The combination of 100 IU / ml rIFN-γ plus 60 IU / ml TNF-α resulted in an additive effect showing a total of 48.7 % inhibition on tachyzoite replication. In contrast, no additive effect was observed when 500 IU / ml rIFN-γ was combined with rTNF-α. No augmentation of rIFN-γ activity was induced by association with rIFN-β. As previously reported 4, 11, in the presence of excess tryptophan (1 mM) the anti-Toxoplasma activity induced by IFN-γ was mostly reversed. In contrast, the inhibitory effect induced by rTNF-α and rIFN-β was only marginally affected by the addition of tryptophan (data not shown).
|
|
T. gondii replicationa) |
|
|---|---|---|
|
Cytokine (IU / ml) |
Infection index |
Inhibition of tachyzoite growth (%) |
|
Control without cytokine |
168.8 ± 27.5 |
− |
|
rIFN-β (1000) |
125.3 ± 9.0 |
25.4 |
|
rTNF-α (60) |
123.5 ± 11.2 |
26.8 |
|
rIFN-γ (100) |
106.7 ± 5.0 |
36.4 |
|
rIFN-γ (100) + rIFN-β (1000) |
103.2 ± 4.8 |
38.6 |
|
rIFN-γ (100) + rTNF-α (60) |
86.2 ± 3.8b) |
48.7 |
|
rIFN-γ (500) |
93.6 ± 16.2 |
43.8 |
|
rIFN-γ (500) + rIFN-β (1000) |
88.9 ± 11.0 |
47.3 |
|
rIFN-γ (500) + rTNF-α (60) |
72.2 ± 19.2 |
56.8 |
- a) Results indicate the means ± SEM from three independent experiments done in duplicate. All results were statistically different (p < 0.05) when comparing experimental and control data.
- b) The difference is statistically significant (p < 0.05) when comparing results from cells activated with 100 IU / ml rIFN-γ plus 60 IU / ml rTNF-α and 100 IU / ml rIFN-γ alone. 2C4 cells were incubated with rIFN-γ, rIFN-β or rTNF-α, and parasite replication evaluated 24 h post-infection.
rIL-4 antagonized the rIFN-γ activity in 2C4 cells, reducing by 67.0 % the inhibitory effect of rIFN-γ on parasite replication. In contrast, pre-incubation with rIL-10 did not interfere with the control of T. gondii replication in human fibroblasts activated with rIFN-γ. No antagonistic effect of rIL-4 or rIL-10 was observed in the rIFN-β, rTNF-α- or rIFN-γ plus rTNF-α-induced inhibition of tachyzoite growth (Table 2).
|
Cytokine |
T. gondii replicationa) |
|
|---|---|---|
|
Infection index |
Inhibition of tachyzoite growth (%) |
|
|
Control without cytokine |
190.9 ± 13.7 |
− |
|
rIL-4 |
184.0 ± 9.3b) |
3.2 |
|
rIL-10 |
184.9 ± 17.9b) |
2.7 |
|
rIFN-γ |
108.3 ± 2.8c) |
43.0 |
|
rIFN-γ + rIL-4 |
163.0 ± 15.9b, d) |
14.2 |
|
rIFN-γ + rIL-10 |
119.3 ± 7.9c) |
37.2 |
|
rIFN-β |
142.9 ± 5.0c) |
24.8 |
|
rIFN-β + rIL-4 |
147.0 ± 6.8c) |
22.6 |
|
rIFNβ + rIL-10 |
145.9 ± 9.0c) |
23.2 |
|
rTNF-α |
137.9 ± 9.4c) |
27.4 |
|
rTNF-α + rIL-4 |
140.6 ± 14.9c) |
26.0 |
|
rTNF-α + rIL-10 |
139.6 ± 15.3c) |
26.4 |
|
rIFN-γ + rTNF-α |
85.7 ± 4.2c) |
54.7 |
|
rIFN-γ + rTNF-α + rIL-4 |
95.2 ± 12.1c) |
49.9 |
- a) Results indicate the means ± SEM from three independent experiments done in duplicate.
- b – d) The differences are either statistically (p < 0.05) not significant (b) or significant (c) when comparing experimental and control data. The difference is statistically significant (d) (p < 0.05) when comparing results from cells activated with rIFN-γ combined with rIL-4 and rIFN-γ alone. 2C4 cells were incubated with 10 ng / ml rIL-4 and 10 ng / ml rIL-10 before and during activation with 100 IU / ml rIFN-γ, 1000 IU / ml rIFN-β or 60 IU / ml rTNF-α, and parasite replication evaluated 24 h post-infection.
2.2 Cytokine control of INDO activity
IFN-γ-induced tryptophan degradation by INDO has been shown to be a major mechanism involved in control of T. gondii replication in human fibroblasts 4, 11, 13. Therefore, we decided to evaluate the effect of different cytokines on tryptophan degradation by INDO. The INDO activity was estimated as the level of tryptophan consumed and kynurenine formed in the culture supernatants of fibroblasts activated with different cytokines. Our results show that the increase of INDO activity induced by different cytokines was inversely correlated with the parasite replication in 2C4 cells. rIFN-γ strongly stimulated INDO activity, with a corresponding fall in tryptophan and elevation of kynurenine levels in the fibroblast supernatants. In contrast to rIFN-γ, rIFN-β and rTNF-α had a weak stimulatory effect on INDO activity. Incubation with rIFN-β (1,000 IU / ml), rTNF-α (60 IU / ml) and rIFN-γ (100 and 500 IU / ml) resulted in 12.8, 29.6, 59.1 and 85.7 % of tryptophan reduction, respectively. Activation of the cells with rIFN-γ (100 IU / ml) plus rTNF-α (but not with rIFN-β) resulted in a statistically significant augmentation of tryptophan degradation (Table 3).
|
Cytokine (IU / ml) |
INDO activitya) |
|||
|---|---|---|---|---|
|
TRP (μM) |
KYN (μM) |
TRP reduction (%) |
KYN formation (μM) |
|
|
Control without cytokine |
18.7 ± 1.2 |
0.7 ± 0.2 |
− |
− |
|
rIFN-β (1000) |
16.3 ± 0.9 |
1.9 ± 0.8 |
12.8 |
1.2 |
|
rTNF-α (60) |
13.2 ± 1.4 |
4.1 ± 0.6 |
29.6 |
3.4 |
|
rIFN-γ (100) |
7.7 ± 1.2 |
7.5 ± 1.0 |
59.1 |
6.8 |
|
rIFN-γ (100) + rIFN-β (1000) |
6.5 ± 1.1 |
8.5 ± 1.3 |
65.5 |
7.8 |
|
rIFN-γ (100) + rTNF-α (60) |
3.2 ± 0.9b) |
11.8 ± 1.1b) |
83.2 |
11.1 |
|
rIFN-γ (500) |
2.7 ± 0.4 |
12.6 ± 0.4 |
85.7 |
11.9 |
|
rIFN-γ (500) + rIFN-β (1000) |
1.8 ± 0.7 |
13.6 ± 0.8 |
90.6 |
12.9 |
|
rIFN-γ (500) + rTNF-α (60) |
1.6 ± 0.6 |
14.6 ± 1.9 |
91.6 |
13.4 |
- a) TRP = tryptophan and KYN = kynurenine. Results indicate the means ± SEM from two independent experiments done in duplicate. All results were statistically different (p < 0.05) when comparing experimental and control data.
- b) The difference is statistically significant (p < 0.05) when comparing results from cells activated with 100 IU / ml rIFN-γ plus 60 IU / ml rTNF-α and 100 IU / ml rIFN-γ alone. 2C4 cells were incubated with rIFN-γ, rIFN-β or rTNF-α, and tryptophan and kynurenine levels evaluated in supernatants 24 h post cytokine stimulation.
Pre-incubation with rIL-4 resulted in 37.6 % inhibition of kynurenine formation and 34.5 % of tryptophan degradation in 2C4 cells activated with rIFN-γ. Similar results were obtained with human lung fibroblasts (data not shown). No reduction of INDO activity by rIL-4 was observed when 2C4 cells were stimulated with either rIFN-β, rTNF-α or with the combination of rIFN-γ plus rTNF-α. Similarly, at higher rIFN-γ concentration (500 IU / ml) the INDO activity was not inhibited by rIL-4 (data not show). In contrast to rIL-4, pre-incubation with rIL-10 did not interfere with the rIFN-β, rTNF-α or rIFN-γ-induced INDO activity in the 2C4 cells (Table 4).
Further, radioactive tryptophan was used to confirm the HPLC results showing the rIL-4 antagonistic effect on INDO activity (Table 4). The activation of 2C4 cells with rIFN-γ resulted in formation of radioactive kynurenine. Thus, the peak of kynurenine, obtained in the HPLC analysis, was indeed generated from tryptophan degradation. In agreement with results obtained with non-radioactive tryptophan, pre-incubation with rIL-4 resulted in 35.0 % inhibition of radioactive formation induced by rIFN-γ (data not shown).
|
Cytokine |
INDO activitya) |
|||
|---|---|---|---|---|
|
TRP (μM) |
KYN (μM) |
TRP reduction (%) |
KYN formation (μM) |
|
|
Control without cytokine |
15.8 ± 1.3 |
0.7 ± 0.3 |
− |
− |
|
rIL-4 |
15.0 ± 1.0b) |
1.9 ± 0.8b) |
4.9 |
1.2 |
|
rIL-10 |
14.8 ± 1.1b) |
1.6 ± 0.6b) |
6.2 |
0.9 |
|
rIFN-γ |
6.0 ± 0.4c) |
9.2 ± 0.8c) |
62.0 |
8.5 |
|
rIFN-γ + rIL-4 |
9.4 ± 0.5c, d) |
6.0 ± 0.3c, d) |
40.6 |
5.3 |
|
rIFN-γ + rIL-10 |
7.0 ± 0.9c) |
9.8 ± 1.3c) |
55.9 |
9.1 |
|
rIFN-β |
13.7 ± 0.9c) |
2.8 ± 0.6c) |
12.9 |
2.1 |
|
rIFN-β + rIL-4 |
14.0 ± 1.1c) |
2.6 ± 0.8c) |
11.4 |
1.9 |
|
rIFN-β + rIL-10 |
13.9 ± 0.8c) |
2.8 ± 1.2c) |
11.8 |
2.1 |
|
rTNF-α |
11.7 ± 0.8c) |
3.7 ± 1.1c) |
25.8 |
3.0 |
|
rTNF-α + rIL-4 |
11.5 ± 0.8c) |
4.1 ± 0.8c) |
27.1 |
3.4 |
|
rTNF-α + rIL-10 |
11.5 ± 0.6c) |
4.0 ± 0.9c) |
27.0 |
3.3 |
|
rIFN-γ + rTNF-α |
2.2 ± 0.3c) |
11.1 ± 1.5c) |
86.1 |
10.4 |
|
rIFN-γ + rTNF-α + rIL-4 |
2.6 ± 0.5c) |
10.5 ± 1.2c) |
84.0 |
9.8 |
- a) TRP = tryptophan and KYN = kynurenine. Results indicate the means ± SEM from two independent experiments done in duplicate.
- b – d) The differences are either statistically (p < 0.05) not significant (b) or significant (c) when comparing experimental and control data. The difference is statistically significant (d) (p < 0.05) when comparing results from cells activated with rIFN-γ combined with rIL-4 and rIFN-γ alone. 2C4 cells were incubated with 10 ng / ml rIL-4 and 10 ng / ml rIL-10 before and during activation with 100 IU / ml rIFN-γ, 1000 IU / ml rIFN-β or 60 IU / ml rTNF-α, and tryptophan and kynurenine levels evaluated in supernatants 24 h post cytokine stimulation.
2.3 Cytokine control of INDO mRNA expression
As shown in Fig. 1, INDO mRNA transcripts were not detectable in untreated fibroblasts. The activation of 2C4 cells with rIFN-β, rTNF-α and rIFN-γ resulted in induction of INDO mRNA. A more intense expression of INDO mRNA was observed in 2C4 fibroblasts activated with rIFN-γ. Incubation of cells with 100 IU / ml rIFN-γ plus 60 IU / ml rTNF-α resulted in an additive effect on INDO mRNA expression (Fig. 1 A and C). The cytokine-induced INDO mRNA expression was measured by densitometric analysis and is shown in Table 1 B and D. As noticed in the INDO activity and control of intracellular tachyzoite replication, rIL-4 decreased the rIFN-γ-induced INDO expression by 53.3 %. A similar inhibitory effect of rIL-4 on INDO mRNA expression was observed in human lung fibroblasts exposed to 100 IU / ml IFN-γ (data not shown). Together, these results suggest that rIL-4 inhibits INDO activity at the transcriptional level. In contrast to rIL-4, rIL-10 was unable to alter the rIFN-γ-induced INDO expression (Fig. 1 C and D).
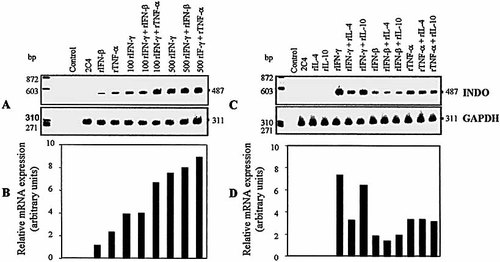
Expression of INDO in 2C4 fibroblasts 14 h after stimulation with rIFN-γ, rIFN-β, rTNF-α, rIL-4 and rIL-10. (A, C) RT-PCR analysis of the amplification product for the INDO gene (487 bp) and the housekeeping gene GAPD (311bp), used as internal control. (B, D) Semi-quantitative RT-PCR analysis of INDO expression. Histograms represent the relative intensity of the transcripts expressed in arbitrary units after standardization on GAPDH level expression. The experiment shown is representative of four separate experiments. bp = base pairs.
2.4 Inhibition of rIFN-γ-induced CD2 and HLA-DR expression by rIL-4
To evaluate the ability of rIL-4 to regulate the expression of other genes induced by IFN-γ, we measured the expression of stably transfected CD2 and endogenous HLA-DR 26, 27 in the 2C4 cell line. The fibroblasts were cultured in the presence or absence of rIFN-γ with or without rIL-4, and the surface expression of such molecules was analyzed be immunostaining with PE-conjugated anti-CD2 and FITC-conjugated anti-HLA-DR mAb. Fig. 2 shows the histograms of a FACS analysis and the percentage of stained 2C4 cells. After treatment with rIFN-γ 59.0 and 16.0 % of the cells became positive for CD2 and HLA-DR antigens, respectively. rIL-4 treatment did not change considerably the percentage of cells expressing CD2 and HLA-DR. In the presence of rIL-4, the rIFN-γ-mediated increase of CD2 and HLA-DR expression was blocked by 17.7 % and 43.8 % (p < 0.05), respectively. In contrast to rIL-4, the addition of rIL-10 showed no effect either on CD2 or HLA-DR expression in the human fibroblast cell line activated with rIFN-γ (data not show).
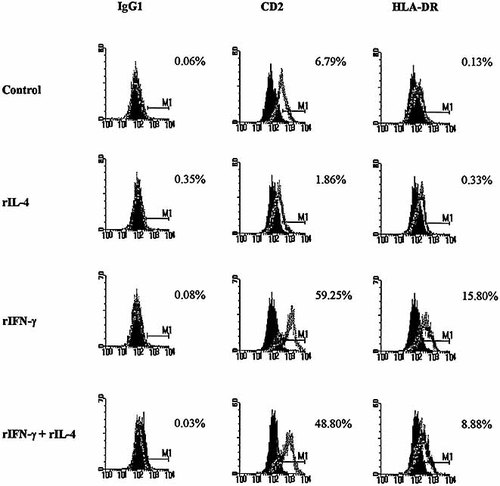
Flow cytometric analysis of CD2 and HLA-DR expression in 2C4 human fibroblasts. The cells were stimulated with 100 IU / ml rIFN-γ and / or 10 ng / ml rIL-4 for 48 h. rIL-4 blocked the IFN-γ-induced CD2 and HLA-DR expression by 17.7 and 43.8 %, respectively (p < 0.05). One representative experiment of two is shown.
2.5 Detection of IL-4, IL-10 and IL-13 receptor mRNA by RT-PCR
IL-4R and IL-13R are multimeric and share at least one common chain called IL-4Rα. Specific IL-13R are formed by the association between either IL-4-Rα and IL-13Rα2 or between IL-13Rα1 and IL-13Rα2. IL-4R, in non-hematopoietic cells, is a heterodimer composed of IL-4Rα and IL-13Rα1 chains 28. To characterize the expression of IL-4R and IL-10R mRNA in human fibroblasts, reverse transcription (RT)-PCR analysis was conducted using sets of primers described in Sect. 4.4. Specific transcripts for IL-4Rα, IL-13Rα1 and IL-13Rα2 chains were detected in 2C4 fibrosarcoma, as well in the cell lines LL-24 and MRC-5 of human lung fibroblasts stimulated or not with different cytokines (Fig. 3 A). Our data show that human fibroblast cells constitutively express the mRNA for different chains of IL-4R and IL-13R. As can be seen in Fig. 3 B, the specific transcript for IL-10R was detected in human monocytic THP-1 cells, but not in the human fibroblast cell lines. The transcript for the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH); used as internal control) was equally expressed in all samples. Together with the functional data, the RT-PCR results suggest that human fibroblasts express specific receptors for IL-4 and IL-13, but not for IL-10.
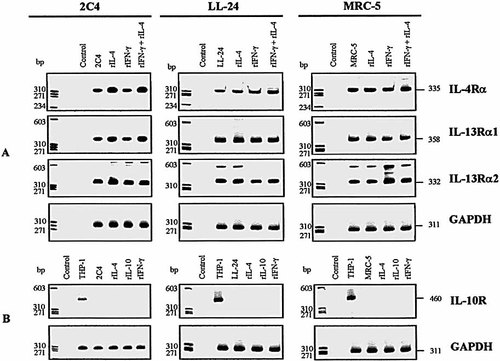
Expression of IL-4R and IL-13R (A) and IL-10R mRNA (B) in 2C4 cells and human lung fibroblasts (LL24 and MRC5) cultured in the presence of rIFN-γ, rIL-4 and/or rIL-10. RT-PCR analysis of the amplification product for the IL-4Rα (335 bp), IL-13Rα1 (358 bp), IL-13Rα2 (332 bp), IL-10R (460 bp) genes as well as the housekeeping gene GAPDH (311 bp), which was used as internal control. Monocyte (THP-1) cDNA was used as positive control in RT-PCR analysis of IL-10R mRNA. bp = base pairs.
2.6 Inhibition of rIFN-γ-induced INDO activity and microbiostasis by IL-13
Since all the fibroblast cell lines were shown to express mRNA for the different chains that compose the IL-13R (Fig. 3 A), we tested the activity of rIL-13 in human fibroblasts. The results shown in Table 5 show that on its own rIL-13 had no effect on INDO activity or parasite replication in the 2C4 cell line. In contrast, rIL-13 inhibited the trytophan degradation (33.0 %), kynurenine formation (41.5 %) and anti-Toxoplasma activity (75.7 %) in IFN-γ-activated fibroblasts. Pre-incubation of IFN-γ-activated fibroblasts with rIL-4, rIL-13 or rIL-4 plus rIL-13 resulted in similar levels of inhibition of INDO activity and microbiostasis.
|
Cytokine |
T. gondii replication |
INDO activity |
||||
|---|---|---|---|---|---|---|
|
|
Infection index |
Inhibition of tachyzoite growth (%) |
TRP (μM) |
KYN (μM) |
TRP Reduction (%) |
KYN Formation (μM) |
|
Control without cytokine |
193.5 ± 7.6 |
− |
18.9 ± 2.1 |
0.2 ± 0.1 |
– |
– |
|
rIL-4 |
182.9 ± 13.7b) |
5.5 |
18.1 ± 1.8b) |
0.4 ± 0.1b) |
4.2 |
0.2 |
|
rIL-13 |
185.4 ± 10.1b) |
4.2 |
17.9 ± 1.2b) |
0.5 ± 0.1b) |
5.3 |
0.3 |
|
rIL-4 + rIL-13 |
180.5 ± 9.5b) |
6.7 |
17.5 ± 2.0b) |
0.7 ± 0.2b) |
7.4 |
0.3 |
|
rIFN-γ |
121.7 ± 2.2c) |
37.1 |
7.9 ± 1.1c) |
8.4 ± 1.1c) |
58.2 |
8.2 |
|
rIFN-γ + rIL-4 |
172.4 ± 2.6c, d) |
10.9 |
11.1 ± 0.9c, d) |
5.2 ± 0.9c, d |
41.3 |
5.0 |
|
rIFN-γ + rIL-13 |
175.9 ± 6.5c, d) |
9.1 |
11.5 ± 1.9c, d) |
5.0 ± 1.2c, d) |
39.0 |
4.8 |
|
rIFN-γ + rIL-4 + rIL-13 |
179.0 ± 8.2c, d) |
7.5 |
11.8 ± 2.4c, d) |
4.7 ± 2.1c, d) |
37.8 |
4.5 |
- a) Results indicate the means ± SEM from two independent experiments done in duplicate.
- b – d) The differences are either statistically (p < 0.05) not significant (b) and significant (c) when comparing experimental and control data. The difference is statistically significant (d) (p < 0.05) when comparing results from cells activated with rIFN-γ combined with rIL-4 and / or rIL-13 and rIFN-γ alone. 2C4 cells were incubated with 10 ng/ ml rIL-4 and / or 10 ng / ml rIL-13 before and during activation with 100 IU / ml rIFN-γ, and tryptophan and kynurenine levels measured in supernatants at 24 h post cytokine stimulation. The parasite replication was evaluated 24 h post-infection.
3 Discussion
Substantial evidence indicates that induction of INDO expression and consequent tryptophan degradation, triggered by cytokines, is largely responsible for the inhibition of intracellular T. gondii replication in human fibroblasts 4, 11, 13, 29 and other NPPC 7, 10, 12, 30. The results presented here confirm that type I and II IFN as well as TNF-α do indeed stimulate INDO expression as well as inhibition of tachyzoite replication within human fibroblast lineage cells. Among the stimulatory cytokines, rIFN-γ was shown to be the most potent inducer of anti-Toxoplasma activity in fibroblasts. Although to a lesser extent, other cytokines such as rIFN-β and rTNF-α also induced significant expression of INDO in human fibroblasts, which resulted in inhibition of T. gondii replication. Furthermore, rTNF-α was shown to increase in an additive manner the induction of INDO expression and activity as well as the inhibitory effect of rIFN-γ on intracellular tachyzoite growth in 2C4 cells. However, the anti-Toxoplasma activity of TNF-α and IFN-β was only marginally reversed by addition of extra tryptophan, suggesting that these two cytokines may trigger another mechanism with relatively minor role in the control of T. gondii replication in human fibroblasts.
IL-4, IL-10 and IL-13 have been frequently shown to display biological activities opposed to those induced by IFN-γ in hematopoietic 17 – 25, 31 – 38 and non-hematopietic 39 – 44 human cells. These studies include the control of IFN-γ-induced INDO expression and activity by IL-4 and IL-10 in cells from monocytic lineage 24. However, no correlation has been established between the inhibitory activity of IL-4 and IL-10 on INDO expression and microbiostatic activity displayed by these cells activated with IFN-γ. Further, the modulatory effect of IL-4 and IL-10 on INDO expression and activity displayed by NPPC stimulated with IFN-γ has not been evaluated. Thus, an important goal of this study was the evaluation of the regulatory role of IL-4, IL-10 and IL-13 on induction of tryptophan degradation and anti-Toxoplasma activity in cells from fibroblast lineage exposed to different types of IFN and / or TNF-α.
As previously reported in cells from monocytic lineage 25, our results indicate that IL-4 partially abrogates the effect of IFN-γ on induction of INDO mRNA expression and tryptophan degradation in human fibroblasts. More precisely, our data show that rIL-4 inhibits the transcription of the INDO gene in 2C4 cells activated with IFN-γ. Consistent with these findings rIL-4 also antagonized the inhibitory effect of IFN-γ on intracellular replication of T. gondii, showing a biological relevance of rIL-4 regulatory activity on tryptophan catabolism. The antagonistic effect of IL-4 upon IFN-γ-induced activities was not restricted to INDO expression. Thus, the rIFN-γ-induced CD2 and HLA-DR expression by 2C4 cells was significantly blocked in the presence of rIL-4.
Many studies have shown that IL-10 antagonizes the expression of various genes and functions induced by IFN-γ in cells from monocytic lineage 17 – 21, 31, 33, including tryptophan degradation 24. Unexpectedly, our data show that rIL-10 had not effect on IFN-γ-induced INDO expression, tryptophan degradation and inhibition of T. gondii replication in 2C4 cells. Similarly, rIL-10 did not interfere with the expression of CD2 and HLA-DR on the surface of 2C4 cells exposed to IFN-γ. In agreement with our findings are studies showing that IL-4 and IL-13, but not IL-10, can inhibit the gene expression induced by IFN-γ in human fibroblasts and other NPPC 39 – 44.
The effects of IL-4 and IL-10 are mediated by their interaction with specific receptors expressed on the surface of a wide variety of cells. However, no information concerning expression of IL-4R and IL-10R on 2C4 cells and IL-10R in human fibroblasts were available. Nevertheless, some studies have demonstrated that human fibroblasts derived from synovial tissue and skin express functional IL-4R and IL-13R 45, 46. Our RT-PCR data demonstrated that human 2C4 fibroblast cells express mRNA for specific chains of IL-4R and IL-13R. In agreement, we also demonstrated the inhibitory effect of rIL-13 on tryptophan degradation and anti-Toxoplasma activity exhibited by IFN-γ-activated fibroblasts. In contrast, we were unable to detect expression of IL-10R mRNA on human fibroblasts. These results are consistent and may explain our findings showing that IL-4 and IL-13, but not IL-10, regulate INDO expression, tryptophan degradation and inhibition of T. gondii replication in 2C4 cells activated by IFN-γ.
Different studies indicate that the antagonistic effect of IL-4 and IL-13 on cellular functions induced by IFN-γ involves the suppression of gene transcription 47. The main signaling pathway triggered by both IL-4 and IL-13 involves phosphorylation of STAT6 47, 48. IFN-γ and IL-4 / IL-13 stimulate the activation of two separate members of the STAT family, STAT1 and STAT6 respectively, both of which recognize the signal-binding element of STAT (SBE) from the interferon-regulatory factor 1 (IRF-1) gene 49. IRF-1 binding to a sequence found in promoters of many IFN-γ-inducible secondary genes 47. Ohmori and Hamilton 50 found that IL-4-induced STAT6 homodimers may effectively compete with homodimeric STAT1 for occupancy of the IRF-1 SBE without providing a positive trans-activating function and may indeed have a negative regulatory function in activation of IFN-γ-inducible genes. It is interesting that rIL-4 had no inhibitory effect on INDO mRNA expression, tryptophan degradation and control of parasite replication induced by TNF-α and IFN-β. This is consistent with the hypothesis that STAT6 may compete with homodimeric STAT1, but not with STAT1 / STAT2 dimer responsible for gene activation induced by IFN-β 51. The lack of rIL-4 inhibition on TNF-α activity could be explained by the fact that TNF-α triggers a distinct signaling pathway not involving activation of members of the STAT family 52. An alternative hypothesis that explains the inhibitory effect of rIL-4 / rIL-13 would be the induction of IRF-2 that competes with IRF-1 for the common binding site in the genomic DNA, thus obstructing IRF-1 from activating gene expression induced by IFN-γ 53. It is noteworthy that IRF-1 is a transcription factor of primary importance for INDO gene expression triggered by IFN-γ 54, 55.
The effects of the down-regulatory cytokines IL-4, IL-10 and TGF-β on microbiostatic / microbicidal activity displayed by PPC from both human 16, 56, 57 and mouse 16 – 21, 32 origin have been studied in detail. A recent study showed that TGF-β also abrogates the IFN-γ-induced expression of INDO mRNA as well as tryptophan degradation 58 in human synovial and skin fibroblasts. However, the effect of TGF-β on microbiostasis displayed by these cells was not evaluated. To our knowledge this is the first study showing the ability of a modulatory cytokine in antagonizing the anti-parasite activity displayed by NPPC activated with IFN-γ. Furthermore, contrasting with studies employing cells from macrophage lineage, our findings indicate that IL-4 and IL-13, but not IL-10, is involved in regulation of INDO expression and activity in cells from fibroblast lineage. Taking into consideration studies performed elsewhere 39, 53 these findings may be extended to other NPPC. Finally, our data argue that prolonged stimulation of the immune system that favors the production of IL-4 and IL-13 over IFN-γ may result in generalized suppression of INDO expression and antimicrobial activity both in PPC and NPPC activated with IFN-γ, thus leading to enhanced host susceptibility to intracellular pathogens 2, 59, 60.
4 Materials and methods
4.1 Cell culture
The human fibrosarcoma 2C4 26, the human lung fibroblast LL-24 (ATCC, CCL-171) and MRC-5 (ATCC, CCL-151) and the human monocytic THP-1 (ATCC, 45502) cell lines were used. The cells were grown in DMEM (Gibco, Grand Island, NY) or RPMI 1640 medium (Gibco) supplemented with 10 % heat-inactivated FBS (Gibco), 5 μM L-glutamine (Sigma, St. Louis, MO), 40 μg / ml gentamicin and 25 mM Hepes buffer, pH 7.3 (Sigma). The tryptophan concentration in the RPMI and the DMEM media was 5 mg / l (24.5 μM) and 16 mg / l (78.4 μM), respectively. The cells were cultured at 37 °C in humidified air containing 5 % CO2. Their viability after trypsin treatment was assessed by trypan blue exclusion.
4.2 Cytokines
Human rIFN-γ, rIFN-β and rTNF-α were provided by Genentech, Inc. (San Francisco, CA) and human rIL-4 and rIL-13 was purchased from R&D Systems, Inc. (Minneapolis, MN). The concentrations used in the assays were 100 and 500 IU / ml for rIFN-γ, 1,000 IU / ml for rIFN-β, 60 IU / ml for rTNF-α and 10 ng / ml for rIL-4, rIL-10 and rIL-13. The appropriate concentrations of each cytokine employed in our study were determined after tritration experiments.
4.3 T. gondii and growth assay
The RH strain of T. gondii was used in the parasite growth assays 4, 11. The parasites were maintained by serial passages in 2C4 human fibroblast monolayer cultures. T. gondii tachyzoite forms were obtained on the second day of the cell culture. Confluent cells were trypsinized (0.034 % trypsin in 0.1 % EDTA, both from Sigma), added to eight-well tissue culture chamber slides (Nunc, Inc.) in duplicates at a final concentration of 3 × 104 cells / well in 430 μl RPMI in the presence or absence of cytokines, and incubated at 37 °C in 5 % CO2 at 95 % humidity for 24 h. The medium was changed after 24 h, and T. gondii was added at a ratio of three parasites per cell in a volume of 200 μl, with or without the cytokines. After 3 h of infection, the parasites that had not infected the cells were removed by washing, replaced with 430 μl fresh medium, and further incubated with or without the cytokines for 24 h. After the incubation, the chambers were washed with PBS, fixed with methanol, stained with May-Grünwald-Giemsa, and mounted on slides. The results were expressed as infection index, i. e. the average number of parasites per 100 infected cells obtained from three independent experiments performed in duplicate.
4.4 mRNA expression assay
To verify the mRNA expression, 24-well culture plates containing 3 × 104 cells / well were used 11. Briefly, the cells were incubated in the presence or absence of cytokines for 14 – 16 h. The medium was discarded, and the cells were washed with PBS. Total RNA was extracted with TRIzol (Gibco) according to the manufacturer's instructions. The cDNA synthesis was obtained in a final volume of 30 μl containing 200 U Moloney murine leukemia virus reverse transcriptase (Pharmacia Biotech, Uppsala, Sweden), 200 mM of each deoxynucleoside triphosphate (Promega, Madison, WI), 2.5 μl buffer (0.1 M MgCl2, 0.5 M Tris-HCl, 1 mM DTT, 2 mg BSA per ml; pH 7.2 at 20 °C, Boehringer Mannheim, Germany), 240 pmol / μl of oligo-dT10 (Boehringer), 0.1 M DTT, 1 U / μl of the RNase inhibitor Rnasin (Promega), and 0.4 μg total RNA. The samples were incubated at 37 °C for 30 min and the cDNA were stored at − 20 °C.
The expression of mRNA was detected in the host cells RT-PCR assays. The human sets of primers were as follows: INDO: forward, 5′-AGT TGA AGT TAA ACA TGC-3prime;; reverse, 5′-TGA TCG TGG ATT TGG TGA-3prime;, amplification product, 487 bp (55 °C, 30 cycles); IL4Rα: forward, 5′-GAC CTG GAG CAA CCC GTA TC-3prime;; reverse, 5′-CAT AGC ACA ACA GGC AGA CG-3prime;, amplification product, 335 bp (60 °C, 30 cycles); IL-13Rα1: forward, 5′-AGG ATG ACA AAC TCT GGA-3prime;; reverse, 5′-CTC AAG GTC ACA GTG AAG G-3prime;, amplification product, 358 bp (55 °C, 30 cycles); IL-13 Rα2: forward 5′-TGG GAC CTA TTC CAG CAA-3prime;; reverse, 5′-TTT TTG GGT AGG TGT TTG GC-3prime;; amplification product, 332 bp (55 °C, 30 cycles); IL-10R: forward 5′-CCA TCT TGC TGA CAA CTT CC-3prime;; reverse, 5′-GTG TCT GAT ACT GTC TTG-GC-3prime;; amplification product, 460 bp (60 °C, 30 cycles); GAPDH: forward, 5′-GTG GTG AAG CAG GCG TCG-3prime;; reverse, 5′-GAC TGA GTG TGG CAG GGA-3prime; amplification product, 311 bp (55 °C, 30 cycles). The amplification was carried out in a thermocycler (PTC 100; MJ Research, Inc., Waltham, MA) in a final volume of 10 μl containing 0.5 U Taq DNA polymerase (Cenbiot, Porto Alegre, RS, Brazil), 200 μM of each deoxynucleoside triphosphate, 1.5 mM MgCl2, 50 mM KCl, and 10 mM Tris-HCl (pH 8.5), together with 10.0 and 1.0 pmol of the others and GAPDH primers, respectively. The expression of the genes was detected by electrophoresis through 6 % nondenaturing polyacrylamide gels and silver staining as previously described 61. For semi-quantitative RT-PCR, amplified products of INDO were quantified using specific bands of the housekeeping gene GAPDH transcripts as reference on densitometric scanning.
4.5 Assay of INDO activity
INDO activity was assessed by both reduction in tryptophan and increase in kynurenine concentration in the supernatants of the culture of cells measured by HPLC 62 2C4 human fibroblasts were plated at a density of 3 × 104 cells / well in 24-well culture plates in 500 μl DMEM. At 24 h, the medium was replaced with 300 μl RPMI 1640 containing 24.5 μM tryptophan, or 300 μl RPMI 1640 plus 1 μCi / ml [5-3H] tryptophan (Specific activity 24 Ci / mmol, Amersham Corp.) in presence or absence of the cytokines. The cells were pre-incubated with rIL-4 and rIL-10 for 3 h and then incubated with rIFN-γ, rIFN-β and rTNF-α. After 24 h of incubation the supernatants were removed and deproteinized with 30 % (w / v) trichloroacetic acid (TCA). The tubes were incubated at 50 °C for 30 min to hydrolyze N-formylkynurenine to form kynurenine. After removal of the protein precipitate by centrifugation (2 min, 10,000 × g), samples were stored at − 20 °C. The levels of tryptophan and kynurenine in the acid-soluble supernatants were analyzed directly by HPLC. In the experiments employing radioactive tryptophan, the radioactivity was detected by both continuous monitoring of effluent by UV spectrophotometry and by collecting fractions corresponding to tryptophan and kynurenine and determining the cpm per fraction by scintillation spectrometry (Beckman LS6000Sc).
4.6 HPLC determination of tryptophan and kynurenine
Total free tryptophan and kynurenine were quantitated by HPLC 63. The chromatography was performed in a Shimadzu LC-10A liquid chromatograph. A Nova-Pak C18 reversed-phase column (4 μm particle size, 3.9 × 150 mm I.D.) was purchased prepacked (Waters, Milford, Massachusetts); a guard-Pak Nova-pak C18 column (Waters) was attached between the injector and the analytical column to extend to life of the analytical column. The column was eluted isocratically at a flow rate of 1.0 ml / min with 1 mM potassium phosphate buffer, pH 4.0, containing 5 % (v / v) of methanol. The absorbance of the column effluent was monitored at 265 nm. For determinations, 100 μl protein-free supernatant was injected onto the column. Kynurenine and tryptophan concentrations in the supernatants were determined based on their peak heights relative to standard solutions obtained in the chromatograms. The standard solutions of kynurenine and tryptophan, purchased from Sigma, were prepared fresh each day at concentrations ranging from 5 to 25 μM of kynurenine and tryptophan in RPMI and submitted to precipitation with 30 % TCA and incubation at 50 °C for 30 min. Chromatography of the standards and the samples was performed under the same conditions.
4.7 Flow cytometric analysis
Expression of HLA-DR and CD2 was assayed by immunofluorescent flow cell cytometry as previously reported 27. Briefly, 2.5 × 105 cells were seeded in a 4-cm petri dish (Nunc, Inc.) and after an overnight incubation, they were prestimulated with rIL-4 for 3 h and then treated with rIFN-γ for 48 h. After incubation the fibroblasts were harvested and stained by successive 30 min incubation at 4 °C with PE-conjugated anti-CD2 and FITC-conjugated anti-HLA-DR mAb (Becton Dickinson). Cells incubated under similar conditions either without or with an isotype-matched control antibody (PE-conjugated anti-IgG1, Diatec, Oslo, Norway) were used as controls. After washing, the cells were pelleted and fixed in 1 % paraformaldehyde. Flow cytometric measurements were performed on a Becton Dickinson FACScan (Becton Dickinson, Mountain View, CA). Data from about 1.0 × 105 cells were obtained and analyzed using a Cell-Quest program (Becton Dickinson). The relative FITC or PE fluorescence intensity was analyzed using a single histogram representation.
4.8 Statistical analysis
Differences between groups were assessed with the Student's t-test. p values of < 0.05 were considered statistically significant.
Acknowledgements
To Dr. C. A. Bonjardim for providing the human fibrosarcoma cell line 2C4 and rIFN-β used in this study and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo a Pesquisa do Estado de Minas Gerais (FAPEMIG) and PAPES / FIOCRUZ by the financial support.
- WILEY-VCH
- WILEY-VCH
- WILEY-VCH




