Expression of 11β-hydroxysteroid dehydrogenase in rat osteoblastic cells: Pre-receptor regulation of glucocorticoid responses in bone
Abstract
11β-hydroxysteroid dehydrogenase (11β-HSD) acts as a pre-receptor signaling mechanism for corticosteroids by regulating the access of active glucocorticoids to both glucocorticoid (GR) and mineralocorticoid receptors (MR). To examine the relationship between endogenous glucocorticoid metabolism and osteoblast function, we have characterized the expression of 11β-HSD isozymes in rat osteosarcoma cells. Analysis of mRNA from ROS 25/1, UMR 106 and ROS 17/2.8 cells revealed transcripts for both 11β-HSD type 1 (11β-HSD1) and type 2 (11β-HSD2) in all three cell lines. However, enzyme activity studies showed only high affinity dehydrogenase activity (inactivation of corticosterone (B) to 11-dehydrocorticosterone (A)), characteristic of 11β-HSD2; conversion of B to A was higher in ROS 25/1> UMR 106 cells>ROS 17/2.8. Although all three cell lines had similar numbers of GR (50,000/cell), glucocorticoid modulation of alkaline phosphatase activity and cell proliferation was only detectable in ROS 17/2.8 cells. Further studies showed that 11β-HSD2 activity in each of the cells was potently stimulated by both A and B, but not by synthetic dexamethasone. This effect was blocked by the 11β-HSD inhibitor, 18β-glycyrrhetinic acid (but not by GR or MR antagonists) suggesting direct, allosteric regulation of 11β-HSD2 activity. These data indicate that in osteosarcoma cells 11β-HSD2 plays a key role in controlling GR-mediated responses; cells with relatively high levels of 11β-HSD2 activity were insensitive to glucocorticoids, whilst cells with low levels showed functional responses to both dexamethasone and B. In addition to the established effects of 11β-HSD2 in protecting MR in the kidney and colon, our data suggest that 11β-HSD2 in bone represents an important pre-receptor mechanism in determining ligand availability to GR. J. Cell. Biochem. 81:453–462, 2001. © 2001 Wiley-Liss, Inc.
The adverse effects of glucocorticoids on the skeletal system have been recognised for many years. Patients with Cushing's syndrome have a 50% prevalence of osteoporosis [Ross and Linch, 1982], comparable to a fracture incidence of 30–50% in patients receiving corticosteroid therapy [Swartz and Dluhy, 1978]. Bone formation is decreased by excess glucocorticoid while bone resorption is enhanced, leading to osteopenia and, ultimately osteoporosis [Lukert and Raisz, 1990]. In vivo this is due, in part, to indirect effects of glucocorticoid on bone cell metabolism, brought about by changes in calcium homeostasis as well as the synthesis of skeletal growth factors and hormones [Lukert and Raisz, 1990; Canalis, 1996]. In vitro glucocorticoids have also been shown to act directly on both bone resorbing (osteoclast) and bone forming (osteoblast) cells. In common with their in vivo actions, glucocorticoids stimulate osteoclastic bone resorption [Gronowicz et al., 1990]. Paradoxically, however, they have also been shown to stimulate osteoclast apoptosis [Dempster et al., 1997]. Glucocorticoid actions on osteoblasts are equally complex and may be either direct, by activating or repressing osteoblast gene expression, or indirect, by altering the expression of growth factors synthesized by osteoblasts themselves [Delany, 1994]. These effects appear to be dependent on the stage of osteoblast differentiation, as well as the dose and duration of exposure to glucocorticoid. Consistent with the reduction in bone formation following corticosteroid exposure, glucocorticoids inhibit both osteoblast proliferation [Canalis, 1983] and the function of mature osteoblasts, decreasing synthesis of α1-collagen [Canalis, 1983; Delany et al., 1995] and osteocalcin [Beresford et al., 1984]. However, glucocorticoids have also been shown to be essential for the differentiation of osteoblast precursors into mature osteoblasts and for the formation of mineralizing bone nodules [Bellows et al., 1990; Leboy et al., 1991; Cheng et al., 1994].
Within a given target cell, corticosteroid hormone action is dependent upon several factors, including the expression of glucocorticoid (GR) and mineralocorticoid receptors (MR). Several reports have suggested that both of these are expressed in osteoblastic cells [Chen et al., 1977; Boivin et al., 1994; Liesegang et al., 1994]. Recent studies by ourselves and others have highlighted the pivotal role of target cell hormone metabolism in modulating the local availability of steroid hormones, including sex hormones [Nawata et al., 1995; Schweikert et al., 1995; Lea et al., 1997; Sasano et al., 1997; Eyre et al., 1998; Janssen et al., 1999] and glucocorticoids [Bland et al., 1999]. With respect to corticosteroid hormone action, two isozymes of 11β-hydroxysteroid dehydrogenase (11β-HSD) interconvert hormonally active cortisol or corticosterone (F or B, respectively) to inactive cortisone or 11-dehydrocorticosterone (E or A, respectively). The type 2 isozyme of 11β-HSD (11β-HSD2) has been shown to co-localize with and protect the MR from illicit occupancy by F or B. In contrast, 11β-HSD1 acts as a reductase facilitating the exposure of GR to F or B [Krozowski and Stewart, 1999]. In this study we have characterized the expression, activity, and function of 11β-HSD isozymes in rat osteosarcoma cells.
MATERIALS AND METHODS
Cell Culture
Rat osteosarcoma cell lines (ROS 25/1, UMR 106, ROS 17/2.8) were maintained in Ham's F-12 medium (Gibco BRL, Paisley, UK) supplemented with 5% fetal calf serum (Gibco BRL). Experimental cultures were grown to confluence in 24-well plates, where indicated cells were incubated for 72 h with various treatments: 100 nM 11-dehydrocorticosterone (A; Sigma, Poole, UK); 100 nM corticosterone (B; Sigma); 100 nM aldosterone (aldo; Sigma); and 100 nM dexamethasone (dex; Sigma). Cells were also incubated with the 11β-HSD inhibitor 18β-glycyrrhetinic acid (5 μM) (glyc. acid; Sigma), the GR antagonist RU38486 (10 μM) (RU486; Roussel Uclaf, Roumainville, France) and the MR antagonist RU38752 (10 μM) (RU752; Roussel Uclaf). All treatments were carried out in medium supplemented with 5% charcoal stripped serum (Advanced Protein Products Ltd., Brierley Hill, UK) following a 1 h incubation in serum free medium.
Reverse Transcription of mRNA
Cells were grown in 80 cm2 flasks to 80–100% confluency and total RNA extracted using RNeasy RNA extraction columns according to manufacturers protocols (Qiagen, Chatsworth, CA). DNA contamination was reduced by the use of QiaShredder columns. RNA was reverse transcribed using a Promega Reverse Transcription System. One microgram of total RNA and 0.5 μg of OligodT(15) was incubated at 70°C for 5 min in a final volume of 10 μl. Primer extension was then performed at 42°C for 60 min, following the addition of 1×reaction buffer, containing 50 mM Tris–HCl (pH 8.3), 50 mM KCl, 10 mM MgCl2, 10 nM dithiothreitol and 0.5 mM Spermidine, with 1 mM of each dNTP, 80 U of RNasin ribonuclease inhibitor and 50 U AMV Reverse Transcriptase (RT; high concentration), in a final volume of 50 μl.
RT-PCR Analysis of 11β-Hydroxysteroid Dehydrogenase (11β-HSD) Isoforms
Aliquots (5 μl) of cDNA from RT reactions were used in polymerase chain reactions (PCR) to amplify specific cDNAs. PCR primers were as follows: rat 11β-HSD1 5′ TCA GAC CAG AAA TGC TCC AG, 3′ AGG AGA TGA TGG CAA TGC TG (PCR fragment size=471 bp); rat 11β-HSD2 5′ GAC TAA TGT GAS CCT CTG GGA G, 3′ TCA GTG CTC GGG GTA GAS GGT G (PCR fragment size 319 bp). Reactions were performed using 1×PCR buffer, containing 50 mM KCl, 10 mM Tris–HCl (pH 9.0), and 0.1% Triton X-100, with 1 mM MgCl2, 0.2 mM of each dNTP and primers at 0.3 μM (11β-HSD1) or 0.2 μM (11β-HSD2) with 1 U Taq DNA polymerase in a final volume of 20 μl. Samples were amplified using an initial denaturation cycle of 95°C for 5 min; 30 cycles of 95°C (1 min), 50°C (1 min) (11β-HSD1), or 60°C (1 min) (11β-HSD2); followed by a final elongation step of 72°C for 7 min. PCR reactions were normalized using primers to amplify β-actin cDNA. PCR reactions were set up as described above using the primers 5′ GTC ACC AAC TGG GAC GAC A and 3′ TGG CCA TCT CTT GCT CGA A. Amplification of samples was performed using an initial denaturation step of 95°C (5 min) followed by 30 cycles of 95°C (1 min), 48°C (1 min), and 72°C (1 min), with a final elongation step of 7 min at 72°C.
Analysis of 11β-Hydroxysteroid Dehydrogenase Activity
Metabolism of B, the principal glucocorticoid in rodents was assessed using a modification of previously described methods [Bland et al., 1999]. Briefly cells were incubated with various concentrations of unlabeled B (10–100 nM) in the presence of tracer amounts (2 nM) of [1,2, 6,7]-3H-B (specific activity 70–100 Ci/mmol, DuPont, Boston) for 5 h. Steroids were extracted from the medium using dichloromethane (5–10 vol) and separated by thin layer chromatography (TLC) using ethanol:chloroform (8:92) as the mobile phase. TLC plates were analyzed using a Bioscan imaging detector and the fractional conversion of B to 11-dehydrocorticosterone (A) calculated. 11-oxo-reductase activity was similarly assessed by incubation of cells with various concentrations of unlabeled A (50–2,000 nM) and tracer amounts of 3H-A (5 nM) (synthesized in house [Bujalska et al., 1997a]) for a period of 5 h. In each case cell monolayers were lysed and total protein concentration assessed using a BioRad protein assay (BioRad, Melville, NY). Results were expressed as pM/h/mg protein. Maximal enzyme activity (VMAX) and substrate affinity (KM) for each enzyme were determined by Lineweaver–Burke analyses.
Analysis of Alkaline Phosphatase Activity
Osteoblastic function was assessed by measurement of alkaline phosphatase activity following an adaptation of a Sigma protocol. Cell monolayers were washed twice with a 0.01% salt solution, followed by a 1 h incubation at 4°C in solubilisation solution (0.01% salt solution+0.1% Triton X-100). Enzyme activity was then assessed at 37°C by the release of p-nitrophenol from p-nitrophenyl during a 15-min incubation step. The reaction was stopped by the addition of 1 M NaOH and liberated p-nitrophenol measured at 405 nm. Total protein concentration of each incubate was assessed using a BioRad protein assay reagent according to manufacturers protocol. Results were expressed as μmol p-nitrophenol produced/h/mg protein.
Steroid Binding Assays
The numbers of GR and MR expressed by rat osteosarcoma cells were assessed using whole cell steroid hormone binding assays as described previously [Bland et al., 1998]. Briefly, confluent cells were trypsinized and then washed twice in PBS. Cell pellets were then resuspended in serum free medium to give a final concentration of 1×107 cells/ml. Aliquots (200 μl) of cell suspension were then incubated for 1 h at 37°C in glass tubes with 3H-dexamethasone (3H-dex) (0.8–50 nM; specific activity 82 Ci/mmol, Amersham, UK). Parallel analyses were also carried out in the presence of a 200-fold excess of unlabeled dex. In these tubes, 3H-dex specifically bound to GR is displaced by the excess unlabeled dex to give a non-specific or background value. This can then be subtracted from total binding (3H-dex alone) to give specific dex binding. Analysis of MR expression was carried out in a similar fashion using 3H-aldosterone (3H-aldo) (0.16–10 nM; specific activity 84.8 Ci/mmol, Amersham, UK), with or without a 200-fold excess of unlabeled aldo. Furthermore, because aldo is also able to bind to GR, additional assay replicates were prepared which included a 200-fold excess of the GR antagonist RU486; any remaining binding was therefore specific for MR. After incubations, cells were washed twice in cold PBS to remove unincorporated dex or aldo, resuspended in 200 μl PBS and bound radioactivity analyzed by scintillation counting. Dex and aldo binding kinetics were determined using Scatchard analyses. Data were linearized by plotting specifically bound hormone divided by free hormone (total minus specifically bound) against specifically bound hormone. The slope of the resulting plot corresponded to the binding affinity value (dissociation constant, KD), whilst the intercept with the x-axis corresponded to the total saturable binding value (maximal binding, BMAX). By using the latter, together with Avogadros constant, it was possible to determine the number of GR or MR/cell.
Analysis of Cell Proliferation
The rate of proliferation of osteosarcoma cells was assessed by measurement of nuclear 3H-thymidine incorporation. Following specific treatments, cells were incubated with 0.5 μCi 3H-thymidine (specific activity 80 Ci/mmol; Amersham) for the last 6 h of culture incubation. Unlabeled thymidine was added for the last 5 min to displace any non-specific uptake of 3H-thymidine. Cells were then washed twice in phosphate buffered saline (PBS), followed by 1 ml of cold 5% trichloroacetic acid (TCA) to precipitate proteins, and left on ice for 20 min. The liquid layer was then removed and drained. An aliquot (250 μl) of 0.1 M sodium hydroxide (NaOH) was added to the cells and left at room temperature for 30 min on a shaker. The resulting solubilized nuclear material was then transferred to 4 ml of scintillant and radioactive counts determined by scintillation counting. Data were reported as mean±standard deviation (SD) of radioactive counts per minute (cpm) (n=4).
Statistics
For all the data sets presented, results represent the mean±SD of quadruplicate values for representative experiments. Each experiment was carried out at least three times with similar observations. KM, VMAX, KD, and BMAX data were determined as mean values±SD from three separate experiments. Statistical analysis was performed using one way ANOVA linked to Tukey–Kramer multiple comparison posttests (Instat version 2.04a computer program, GraphPad Software, Inc., San Diego, CA).
RESULTS
Expression and Activity of 11β-HSD in Rat Osteosarcoma Cells
Initial studies were carried out to characterize the expression and activity of 11β-HSD isozymes and corticosteroid receptors in ROS 25/1, UMR 106, and ROS 17/2.8 cells. RT-PCR analysis of mRNA demonstrated the presence of transcripts for both 11β-HSD1 and 11β-HSD2 in all three cell lines (Fig. 1A). However, enzyme assays using intact cells showed only high affinity dehydrogenase activity (i.e., oxidative conversion of B to A) (Fig. 1B). Kinetic analysis of activity data for ROS 25/1 indicated a mean KM value of 25 nM and a VMAX of 14.4 pM/h/mg protein. Corresponding values for UMR 106 were 33.6 nM and 5.5 pM/h/mg protein, respectively. The lowest VMAX was observed with ROS 17/2.8 cells (1.8 pM/h/mg protein) which had a KM of 22 nM. No conversion of A to B was observed in any of the three cell lines (data not shown).
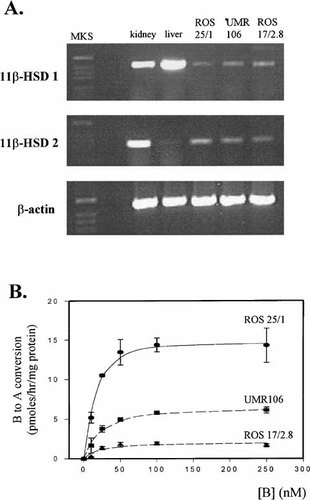
Characterization of 11β-HSD isozyme expression and activity in rat osteosarcoma cells. A: RT-PCR analysis of 11β-HSD1 and 2 in ROS 25/1, UMR 106 and ROS 17/2.8 cells. +ve control RNA: rat liver (11β-HSD1); rat kidney (11β-HSD2). B: Dose–reponsive conversion of B to A using intact cell preparations of ROS 25/1, UMR 106 and ROS 17/2.8.
Corticosteroid Receptor Expression in Rat Osteosarcoma Cells
Binding studies using 3H-dex and 3H-aldo indicated that both GR and MR were expressed in the rat osteosarcoma cells. Data in Figure 2 show typical Scatchard plot analyses that were used to determine maximal binding (GR or MR/cell) and binding affinity (KD) values for dex. Replicate assays (n=3) indicated that there was no significant difference in the mean numbers of GR/cell for each cell line: ROS 25/1, 39,000; UMR 106, 42,200; ROS 17/2.8, 48,000. However, mean KM values for GR in ROS 25/1 (2.8 nM) and UMR 106 (4.1 nM) were lower than in ROS 17/2.8 cells (17.7 nM), indicating lower binding affinity for dex in the latter. Mean KD values for aldo binding were similar for all three cell lines being of the order of 0.4 nM, although ROS 25/1 cells expressed higher numbers of MR/cell (7,500) compared to either UMR 106 (2,500) or ROS 17/2.8 cells (1,600).
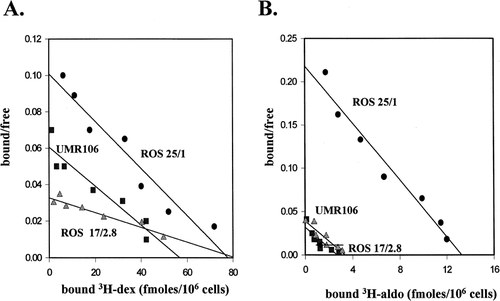
Expression of corticosteroid receptors in rat osteosarcoma cells. A: Scatchard plots for specific binding of radiolabeled dexamethasone (3H-dex) in ROS 25/1, UMR 106, and ROS 17/2.8 cells. B: Scatchard plots for specific binding of radiolabeled aldosterone (3H-aldo) in ROS 25/1, UMR 106, and ROS 17/2.8 cells.
Analysis of Functional Responses to Corticosteroids in Rat Osteosarcoma Cells
Further studies were carried out to assess the impact of 11β-HSD2 activity and MR/GR expression on corticosteroid-induced responses in rat osteosarcoma cells. The effects of A, B, dex, and aldo on osteoblast activity were initially determined using changes in alkaline phosphatase activity as a marker of cell function (Fig. 3). Glucocorticoid-induction of alkaline phosphatase activity was only demonstrable in ROS 17/2.8 cells, with dex showing significantly higher induction than B. Treatment with aldo and A had no effect on alkaline phosphatase activity in any of the cell lines. Parallel studies were carried out to determine the receptor target for glucocorticoid-induced effects on ROS 17/2.8 cells. The GR antagonist RU486 completely inhibited B and dex-induced alkaline phosphatase activity in ROS 17/2.8 cells (Fig. 4). The MR antagonist RU752 also attenuated the induction of alkaline phosphatase activity by B and dex, although the effect was not as striking as that observed with RU486. Treatment for 72 h with 100 nM aldo had no effect on any of the cells in the presence or absence of RU486 or RU752 (data not shown).
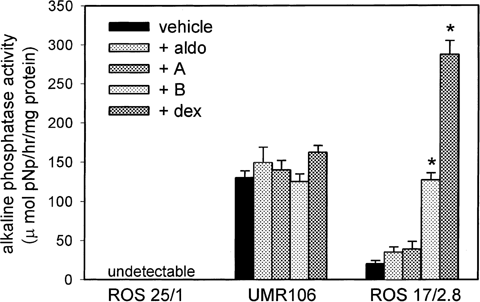
Corticosteroid-mediated regulation of alkaline phosphatase activity in rat osteosarcoma cells. Effect treatment with 100 nM aldosterone (aldo), 100 nM corticosterone (B), 100 nM dehydrocortciosterone (A) or 100 nM dexamethasone (dex) on alkaline phosphatase activity in ROS 25/1, UMR 106 and ROS 17/2.8. All treatments were for 72 h in medium supplemeted with 5% charcoal-stripped FCS. Values are the mean±standard deviation (SD) for n=4 determinations. *, significantly different from untreated ROS 17/2.8, P<0.01.
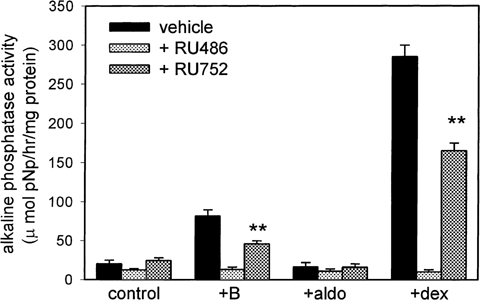
Role of corticosteroid receptors in glucocorticoid-induced regualtion of alkaline phosphatase activity in ROS 17/2.8 cells. Effect of 100 nM B, 100 nM aldosterone (aldo) or 100 nM dexamethasone (dex) on alkaline phosphatase activity in ROS 17/2.8 cells in the presence or absence of 10 μM RU486 or 10 μM RU752. All treatments were for 72 h in medium supplemeted with 5% charcoal-stripped FCS. Values are the mean (±SD) for n=4 determinations. *, sigificantly different from vehicle, P<0.01.
These data suggested that, unlike ROS 17/2.8, cells with relatively high levels of 11β-HSD2 activity such as ROS 25/1 and UMR 106 were unresponsive to glucocorticoids. However, in view of the elevated levels of alkaline phosphatase activity in untreated UMR 106 cells, a further marker of osteoblast status was assessed. Results in Figure 5 show the effect of dex or B (both at 100 nM) on proliferation of ROS 25/1, UMR 106, and ROS 17/2.8 cells as determined by nuclear 3H-thymidine incorporation at 72 h. Whilst dex inhibited proliferation in all three cell lines, only ROS 17/2.8 cells showed significant response to treatment with B.

Glucocorticoids and osteosarcoma cell proliferation. Effect of treatment with 100 nM B or 100 nM dexamethasone (dex) on nuclear 3H-thymidine incorporation in ROS 25/1, UMR 106 and ROS 17/2.8 cells. All treatments were for 72 h in medium supplemented with 5% charcoal-stripped FCS. Values are the mean±standard deviation (SD) for n=4 determinations. *, significantly different from untreated control, P<0.05. **, significantly different from untreated control, P<0.05.
Regulation of 11β-HSD2 Activity in Rat Osteosarcoma Cells
Data in Figure 6 indicated that pre-treatment with either the substrate or product of 11β-HSD2 enhanced the activity of this enzyme. This effect was observed with each of the cell lines and was inhibited by co-treatment with the 11β-HSD inhibitor 18β-glycyrrhetinic acid. In contrast, the GR antagonist RU486 and MR antagonist RU752 were without effect. The synthetic glucocorticoid dex had no significant effect on 11β-HSD2 activity.
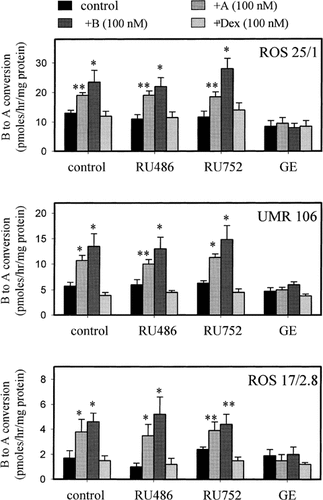
Regulation of 11β-HSD activity in rat osteosarcoma cells. ROS 25/1, UMR 106, and ROS 17/2.8 cells were treated with dexamethasone (dex), corticosterone (A) or 11-dehydrocorticosterone (B) (all at 100 nM) in medium containing 5% charcoal-stripped FCS. Treatments were carried out in the presence or absence of the GR antagonist RU486 (10 μM), the MR antagonist RU752 (10 μM) or the enzyme inhibitor 18β-glycyrrhetinic acid (GE) (5 μM). After incubation for 24 h, growth medium was removed, cell monolayers washed twice with serum-free medium and then incubated for a further 2 h in serum-free medium. Metabolism of B to A was then assessed using conventional methods described above. Values are the mean±standard deviation (SD) for n=4 determinations. *, significantly different from untreated control, P<0.01. **, significantly different from untreated control, P<0.05.
DISCUSSION
The regulation of glucocorticoid levels in target cells is, in part, determined by the enzyme 11β-HSD which catalyzes the interconversion of active 11-hydroxysteroids (F and B) to their inactive metabolites (E and A) [Krozowski and Stewart, 1999]. To date, two isozymes of 11β-HSD have been cloned and characterised. 11β-HSD1 is a low affinity, bi-directional, NADP(H)-dependent enzyme which in vivo acts primarily as a reductase, converting inactive A (or E) to physiologically active B (or F) [Lakshmi and Monder, 1988; Agarwal et al., 1989; Monder and Lakshmi, 1989]. Expression of mRNA for 11β-HSD1 has been demonstrated in several tissues, principally those with high levels of GR but low levels of MR, suggesting that this isozyme serves to modulate the access of glucocorticoids to the GR [Krozowski and Stewart, 1999]. In contrast, 11β-HSD2 is a unidirectional, high affinity, NAD-dependent dehydrogenase [Albiston et al., 1994; Zhou et al., 1995]. 11β-HSD2 is found predominantly in mineralocorticoid target tissues where it serves to protect the MR from non-selective occupation by glucocorticoids [Edwards et al., 1988; Funder et al., 1988]. Mutations in the 11β-HSD2 gene results in the hypersensitive syndrome of apparent mineralocorticoid excess, in which cortisol acts as a potent mineralocorticoid (AME) [Wilson et al., 1995; Stewart et al., 1996]. A recognised but unexplained feature of this syndrome is severe osteopenia [Shimojo and Stewart, 1995].
Using human osteosarcoma cell lines we have recently described the presence of oxidative 11β-HSD activity in osteoblastic cells. This correlated directly with the level of GR expression (MR expression was undetectable in all cell lines) [Bland et al., 1998]. Enzyme kinetics and mRNA analyses suggested that the 11β-HSD activity in human osteosarcoma cells was due to expression of the type 2 isozyme. This was in contrast to analysis of glucocorticoid metabolism in primary cultures of human osteoblasts, which identified mRNA for 11β-HSD1 [Bland et al., 1998]. However, the predominant activity in these cells remained inactivation of F to E. These observations are further supported by data presented in this study using rat osteosarcoma cells. Glucocorticoid metabolism in ROS 25/1, UMR 106, and ROS 17/2.8 cells was exclusively oxidative, in this case conversion of B to A. The kinetics of this enzyme activity were indicative of a high affinity dehydrogenase (KM<40 nM B) which, together with the RT-PCR analyses is consistent with the expression of 11β-HSD2 [Brown et al., 1993]. Although KM values for the putative 11β-HSD2 were similar for each of the rat cell lines, there were consistent and significant differences in maximal enzyme activity (VMAX). Notably, the highest levels of B to A conversion were observed in relatively immature fibroblast-like ROS 25/1 cells, with almost undetectable 11β-HSD2 activity in more mature osteoblastic ROS 17/2.8 cells. These observations are in direct contrast to recent studies of estrogen metabolism in rat osteosarcoma cells in which we showed that both aromatase and 17β-hydroxysteroid dehydrogenase activities were highest in ROS 17/2.8 cells [Eyre et al., 1998].
A further similarity with human osteoblastic cells was the predominant expression of GR in rat osteosarcoma cells. This suggests that, in osteoblastic cells, the function of 11β-HSD2 is to attenuate GR-mediated responses rather than protecting against illicit MR occupancy. However, it is important to recognise that glucocorticoids such as F or B have a much higher affinity for the MR (KD=1 nM) than the GR (KD=10 nM) [Krozowski and Stewart, 1999]. Thus, although numbers of MR are relatively low in rat osteosarcoma cells, receptor transactivation following binding of B to the MR remains a possibility. Indeed, although the induction of alkaline phosphatase activity by B and dex was inhibited completely by the GR antagonist RU486, there was also partial attenuation of this response in the presence of the MR antagonist RU752. The latter occurred even though aldo itself had no effect on alkaline phosphatase activity in ROS 17/2.8 cells. Thus, glucocorticoid responses in ROS 17/2.8 cells may not be mediated exclusively via a classical GR signaling pathway. Although GR are the predominant corticosteroid receptors in bone, it is still possible that low levels of high affinity MR participate in glucocorticoid-induced osteoblast responses, particularly as previous studies have demonstrated the capacity for functional interaction between GR and MR at the level of heterodimer formation [Liu et al., 1995].
The close relationship between 11β-HSD2 and GR in rat osteosarcoma cells confirms previous data from human osteoblastic cells [Bland et al., 1998], and further highlights a possible novel function for this isozyme in bone. It is interesting to contrast these observations with studies of other adult tissues which have suggested that the key function of 11β-HSD2 is to prevent illicit occupancy of MR by glucocorticoids [Edwards et al., 1988; Funder et al., 1988]. Likewise, studies using liver [Jamieson et al., 1995], adipose tissue [Bujalska et al., 1997b; Bujalska et al., 1999] and CNS tissue [Rajan et al., 1996] have demonstrated an association between GR expression and 11β-HSD1, with the latter modulating GR-mediated effects by controlling the local availability of active glucocorticoids. The pattern of glucocorticoid metabolism and corticosteroid receptor expression in osteoblastic cells is therefore clearly different to that observed in most other adult tissues. Indeed, in previous studies we have shown that the association between 11β-HSD2 and GR is more characteristic of fetal tissues, with fetal bone providing a particularly good example [Condon et al., 1998]. The precise function of 11β-HSD2 in fetal tissues remains to be determined. However, data from osteoblastic cells suggest that in some instances the enzyme may act by protecting GR from over-exposure to active glucocorticoids.
The most important aspect of the current study is that although all three osteosarcoma cell lines expressed similar numbers of GR, there were clear differences in terms of their functional responses to glucocorticoids. Cell proliferation data suggested that the responsiveness of ROS 17/2.8 cells to naturally occurring glucocorticoids (such as B) correlated with relatively low levels of 11β-HSD2 activity. Conversely, cells with a higher capacity for glucocorticoid inactivation such as ROS 25/1 or UMR 106 showed antiproliferative responses to synthetic dex but not B. Previous data have shown that dex is poorly metabolized by 11β-HSD2, having a relatively low affinity for the enzyme; the KM for dex in rats is ≈3 μM [Li et al., 1997], and in humans 900 nM [Ferrari et al., 1996]. As such it is unlikely to be subject to significant local metabolism in osteoblastic cells. It was therefore interesting to note that although ROS 25/1 and UMR 106 cells showed anti-proliferative responses to dex, similar doses of the compound had no effect on alkaline phosphatase induction in these cells. The precise explanation for this remains unclear, although in the case of UMR 106 this is most probably due to the relatively high basal levels of alkaline phosphatase expression. Further studies using UMR 106 cells have shown that dex also stimulates expression of mRNA for osteopontin, although (as with the proliferation data) similar doses of corticosterone were without effect (data not shown).
The lack of metabolism of dex by 11β-HSD also raises important questions concerning the physiological relevance of this compared to naturally occurring glucocorticoids, particularly as dex is routinely used as a differentiating agent for in vitro studies of bone. Furthermore, the stimulation of 11β-HSD2 activity following pre-treatment with B or A suggests that it may be difficult to reverse resistance to naturally occurring glucocorticoids in cells expressing relatively high levels of 11β-HSD2. Even at doses as high as 1 μM we were unable to detect significant responses to B in ROS 25/1 or UMR 106 cells, and this may be due to direct stimulation of 11β-HSD2 activity by the corticosteroid itself. In view of the fact that ‘self-regulation‘ is blocked by an HSD inhibitor but not by MR or GR antagonists, it is possible to speculate that the enzyme is subject to allosteric regulation. Previous studies in vivo using mouse kidney tissue have also highlighted the induction of 11β-HSD2 activity by corticosteroids, although in this case the authors postulated that this was due to activation of latent enzyme activity [Li et al., 1996]. Further characterization of this mechanism in osteoblastic cells (particularly with respect to therapeutic corticosteroids such as prednisolone) may help to clarify the apparent dichotomy of responses to glucocorticoids observed in bone.
In summary, the data presented in this study provide evidence of a functional role for local 11β-HSD activity in determining the effects of glucocorticoids on osteosarcoma cells. In contrast to the many target tissues studied to date, expression of the 11β-HSD2 isozyme appears to modulate glucocorticoid exposure to GR rather than MR. As such, regulation of 11β-HSD2 is likely to be a key determinant of both the physiological and pathophysiological effects of glucocorticoids on bone.
Acknowledgements
We thank Drs. G. Rodan (Merck, West Point) and J. Martin (Melbourne) for generously donating the ROS and UMR cell lines, respectively. This work was supported by The United Birmingham Hospitals Endowment Fund and by the Medical Research Council (P.M.S., E.H.R. and M.S.C.).