Silver syndrome is not linked to any of the previously established autosomal dominant hereditary spastic paraplegia loci
Abstract
The hereditary spastic paraplegias are a clinically variable and genetically heterogeneous group of disorders characterized by progressive and lower limb spasticity and weakness. Silver syndrome (SS) is a particularly disabling autosomal dominant form of the disease in which there is associated wasting of the hand muscles. In view of the fact that genes for hereditary spastic paraplegia can produce highly variable phenotypes, the eight known autosomal dominant loci were investigated for linkage to Silver syndrome. Genotyping of these loci in two large multigenerational families was incompatible with linkage to any of these regions, suggesting that an additional locus is responsible for this syndrome. © 2001 Wiley-Liss, Inc.
INTRODUCTION
The hereditary spastic paraplegias (HSPs) are a clinically variable group of disorders comprising progressive lower limb spasticity and weakness. These disorders are classified as ‘pure’ when spasticity occurs in isolation, and ‘complicated’ when additional neurological manifestations, such as mental retardation, deafness and optic neuropathy are present. In addition to the marked clinical variability, HSP is also genetically heterogeneous with eight autosomal dominant, four autosomal recessive and three X-linked forms of the disease mapped [Hazan et al., 1993, 1994; Hentati et al., 1994; Jouet et al., 1994; Saugier-Veber et al., 1994; Fink et al., 1995; Casari et al., 1998; Hedera et al., 1999; Martinez Murrilo et al., 1999; Reid et al., 1999, 2000; Seri et al., 1999; Claes et al., 2000; Fontaine et al., 2000; Vazza et al., 2000]. Although the precise pathogenesis underlying HSP remains uncertain, the principal neuropathological finding seems to be axonal degeneration in the terminal portions of the long descending and ascending tracts. Age-of-onset is variable but the first clinical signs of this disease are typically apparent by the second or third decade of life. The rate of progression and severity of axonal degeneration also display inter and intra-familial variation.
With an estimated prevalence of approximately 1:10,000 HSP has been increasingly recognized as an important hereditary neurodegenerative disorder [Polo et al., 1991]. A particularly disabling and rare form of HSP involving wasting of the hand muscles was originally described by Silver [1966]. This form of HSP, termed Silver syndrome (SS), is inherited in an autosomal dominant fashion and displays an earlier age of onset than many other forms of HSP. Clinically, SS is characterized by marked amyotrophy and weakness of the small muscles of the hands and feet in conjunction with lower limb spasticity [Silver, 1966]. Impairment of distal vibration sense is also seen [Silver, 1966].
Previous studies have shown no clear clinical distinction between any of the autosomal dominant HSP loci [Heinzlef et al., 1998]. Indeed, families in which pure and complicated forms of HSP are segregating have been assigned to the chromosome 2p locus [Heinzlef et al., 1998]. Furthermore the two X-linked conditions, caused by mutations within the L1 cell adhesion molecule (L1-CAM) and proteolipid protein (PLP) genes, have both been associated with pure and complicated forms of HSP. In addition to HSP, mutations within L1-CAM have also been associated with MASA syndrome (mental retardation, aphasia, shuffling gait and adducted thumbs) and X-linked hydrocephalus [Jouet et al., 1994], whereas mutations within the PLP gene have been shown to result in Pelizaeus-Merzbacher disease [Saugier-Veber et al., 1994].
In view of the fact that genes responsible for HSP can produce highly variable phenotypes, we investigated two large multigenerational Caucasian SS families from the UK to determine if this variant of HSP was linked to any of the eight known autosomal dominant loci. Two kindreds with SS were studied. Family 1 is an extended version of that first described 34 years ago [Silver, 1966] whereas Family 2 has not previously been described (Fig. 1).
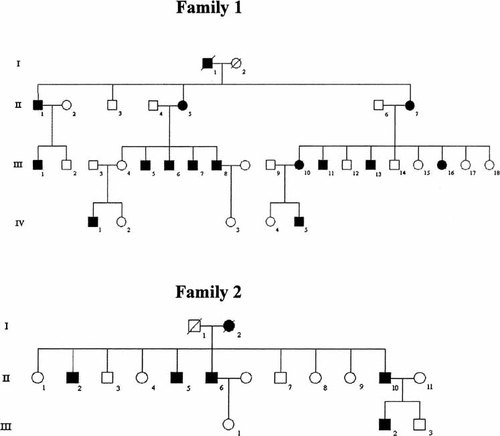
Pedigrees of Families 1 and 2 used to perform linkage analysis.
Clinical evaluation was undertaken by two examiners (Table I) and showed lower limb spasticity and wasting of intrinsic hand muscles in the majority of affected individuals. All intrinsic hand muscles were weak, with severe wasting most marked in the thenar eminence. There were no abnormal cranial nerve signs, including funduscopy, observed. Age of onset was earlier than many other forms of HSP, typically appearing before the age of 17 [Silver, 1966]. Two individuals in Family 1 displayed signs of the disease before their teens (III-4 and III-9). There was also impairment of vibration sense in the lower limbs of older individuals.
| Patient | Family 1 | Family 2 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| II–1 | II–5 | II–7 | III–1 | III–3 | III–5 | III–6 | III–8 | III–11 | III–13 | III–16 | II–6 | II–10 | III–2 | |
| Age at examination | 75 | 77 | 81 | 45 | 18 | 49 | 53 | 55 | 57 | 61 | 42 | 64 | 52 | 23 |
| Age at onset of spastic paraplegia | 25 | 40s | 20s | 15 | 20s | 8 | 28 | 40 | 35 | 7 | ||||
| Sphincter involvement | ++ | + | ++ | |||||||||||
| Pes cavus | ++ | ++ | + | + | − | + | + | − | − | + | − | − | + | |
| Wasting of hands | + | + | ++ | − | − | + | + | + | − | + | ++ | − | ++ | ++ |
| Upper limb pyramidal signs | + | + | − | + | − | − | − | − | − | + | − | − | − | − |
| Lower limb pyramidal signs | ++ | +++ | ++ | + | + | + | ++ | ++ | − | + | + | ++ | + | ++ |
| Sensation | N | N | N | N | N | N | N | N | Vibration loss to ankle | N | N | Vibration loss to ankles | N | |
- * Data for affected family 1 individuals III–7 and IV–1, and family 2 individuals II–2 and II–5 were not available. N, normal; −, absent; +, mild; ++, moderate; +++, severe.
Markers located within and flanking the previously established HSP critical regions of chromosomes 2p21–p22, 2q24–q34, 8q24, 10q23.3–q24.1, 12q13, 14q11.2–q24.3, 15q11.1, and 19q13 were examined for linkage to the disease phenotype using 27 individuals from Family 1 and 14 individuals from Family 2. Multipoint LOD scores were calculated using FastMap program version 1.1 [Curtis and Gurling, 1993].
RESULTS AND DISCUSSION
Linkage analysis of combined data for Families 1 and 2 produced significantly negative LOD scores of less than −2 spanning each locus, excluding linkage to these regions (Fig. 2). The two-point LOD scores generated for each family for markers flanking and within each of the HSP loci are given in Table II. These data demonstrate further locus heterogeneity for AD HSP and suggest that the unique phenotype of SS is caused by another, as yet unmapped, gene. We have now commenced a genome-wide screen for linkage to map the position of this variant of HSP. The molecular pathogenesis of the various forms of HSP is not yet understood. The cloning of the paraplegin gene, which is mutated to cause an autosomal recessive form of HSP, however, has suggested a mitochondrial-based mechanism for this form of the disorder [Casari et al., 1998]. Impairment of mitochondrial function may be a common mechanism underlying HSPs, and an increasing body of evidence implicates this process in a number of neurodegenerative conditions [Schulz et al., 1997]. Recently, the gene responsible for the chromosome 2p-linked form of HSP has been cloned. This gene, termed spastin, is thought to encode a molecule that participates in non-mitochondrial protein complex assembly [Hazan et al., 1999]. Although being only distantly related, both paraplegin and spastin belong to the AAA family of proteins that are thought to act as chaperone molecules. This finding suggests that chaperone protein function may be critical for corticospinal tract preservation.
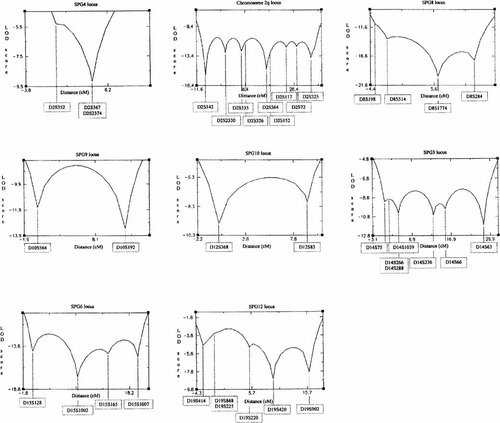
Multipoint linkage maps of combined data for Families 1 and 2 for the candidate HSP loci on chromosomes 2p, 2q, 8q, 10q, 12q, 14q, 15q and 19q. Produced using FastMap [Curtis and Gurling, 1993].
| Marker | Family | LOD score | |||||
|---|---|---|---|---|---|---|---|
| θ = 0.01 | θ = 0.05 | θ = 0.1 | θ = 0.2 | θ = 0.3 | θ = 0.4 | ||
| D2S352 | 2 | −3.32 | −1.37 | −0.64 | −0.12 | 0.02 | 0.02 |
| D2S367 | 1 | −4.05 | −0.86 | 0.24 | 0.88 | 0.79 | 0.36 |
| D2S2374 | 1 | −4.90 | −2.19 | −1.13 | −0.28 | −0.01 | 0.02 |
| 2 | −3.03 | −1.09 | −0.40 | 0.05 | 0.10 | 0.03 | |
| D2S119 | 2 | −1.40 | −0.72 | −0.44 | −0.19 | −0.07 | −0.02 |
| D2S142 | 1 | −4.22 | −2.02 | −1.09 | −0.30 | −0.03 | 0.02 |
| 2 | −3.64 | −2.00 | −1.22 | −0.484 | −0.16 | −0.03 | |
| D2S2330 | 1 | −9.63 | −5.76 | −3.81 | −1.81 | −0.76 | −0.21 |
| D2S364 | 1 | −9.25 | −5.51 | −3.63 | −1.73 | −0.74 | −0.22 |
| D2S72 | 2 | −1.28 | −0.48 | −0.14 | 0.09 | 0.10 | 0.04 |
| D2S325 | 1 | −8.25 | −5.34 | −3.72 | −1.94 | −0.94 | −0.34 |
| D8S198 | 2 | −3.33 | −1.37 | −0.65 | −0.15 | −0.02 | −0.01 |
| D8S514 | 1 | −5.76 | −2.42 | −1.14 | −0.14 | 0.13 | 1.10 |
| D8S1774 | 1 | −13.73 | −6.96 | −4.23 | −1.83 | −0.75 | −0.24 |
| D8S284 | 1 | −6.13 | −2.81 | −1.54 | −0.54 | −0.21 | −0.09 |
| 2 | −5.02 | −2.37 | −1.35 | −0.52 | −0.19 | −0.04 | |
| D10S564 | 1 | −8.67 | −4.01 | −2.20 | −0.73 | −0.18 | −0.02 |
| D10S192 | 1 | −6.72 | −3.35 | −2.00 | −0.84 | −0.31 | −0.07 |
| 2 | −3.79 | −1.80 | −1.03 | −0.40 | −0.14 | −0.03 | |
| D12S368 | 1 | −4.35 | −2.30 | −1.28 | −0.40 | −0.11 | −0.05 |
| D12S83 | 1 | −2.62 | −1.44 | −0.79 | −0.17 | 0.06 | 0.10 |
| 2 | −1.18 | −0.42 | −0.13 | 0.03 | 0.03 | 0.01 | |
| D14S75 | 1 | −4.63 | −2.43 | −1.49 | −0.64 | −0.25 | −0.06 |
| 2 | −2.22 | −0.91 | −0.42 | −0.07 | 0.02 | 0.01 | |
| D14S276 | 1 | −6.54 | −3.17 | −1.83 | −0.68 | −0.19 | −0.01 |
| D14S66 | 2 | −3.03 | −1.09 | −0.40 | 0.05 | 0.10 | 0.03 |
| D14S63 | 1 | −7.82 | −3.20 | −1.44 | −0.10 | 0.28 | 0.21 |
| 2 | −1.72 | −1.04 | −0.75 | −0.44 | −0.20 | −0.05 | |
| D15S128 | 1 | −5.54 | −2.26 | −1.03 | −0.11 | 0.13 | 0.10 |
| 2 | −5.03 | −2.37 | −1.35 | −0.52 | −0.19 | −0.05 | |
| D15S1002 | 1 | −11.44 | −5.40 | −3.01 | −1.00 | −0.18 | 0.09 |
| D15S165 | 2 | −3.40 | −1.87 | −1.10 | −0.39 | −0.12 | −0.02 |
| D15S1007 | 1 | −10.63 | −5.26 | −3.12 | −1.28 | −0.48 | −0.11 |
| D19S414 | 1 | −6.55 | −3.89 | −2.53 | −1.15 | −0.47 | −0.14 |
| D19S225 | 2 | −1.56 | −1.19 | −0.85 | −0.41 | −0.17 | −0.04 |
| D19S220 | 1 | −2.01 | −1.23 | −0.87 | −0.50 | −0.30 | −0.15 |
| D19S902 | 1 | −3.31 | −1.55 | −0.70 | 0.02 | 0.17 | 0.08 |
| 2 | −0.73 | −0.61 | −0.48 | −0.27 | −0.12 | −0.03 | |
- * As markers were not always informative in both families, alternative markers were genotyped for exclusion of linkage to the HSP loci.
The mapping and ultimate cloning of the gene responsible for SS may provide further insight into the underlying mechanisms responsible for these diseases. In turn this will facilitate the identification of the additional genes responsible for other autosomal dominant forms of HSP.




