Genes Involved in Human Ribosome Biogenesis are Transcriptionally Upregulated in Colorectal Cancer
Abstract
Microarray gene expression profiling comprising 168 colorectal adenocarcinomas and 10 normal mucosas showed that over 79% of the genes involved in human ribosome biogenesis are significantly upregulated (log2 > 0.5, p < 10−3) when compared to normal mucosa. Overexpression was independent of microsatellite status. The promoters of the genes studied showed a significant enrichment for several transcription factor binding sites. There was a significant correlation between the number of binding site targets for these transcription factors and the observed gene transcript upregulation. The upregulation of rRNA processing genes points towards a coordinated process enabling the overproduction of matured ribosomal structures.
1. Introduction
Protein translation and ribosome biogenesis are essential cellular processes tightly regulated at different levels. Ribosome biogenesis processes and assembles precursor rRNA into mature rRNA that together with ribosomal proteins become the mature ribosome. Single components of this machinery are deregulated in cancer. Increased cellular growth or proliferation needs an enhanced protein content and protein synthesis [1].
In colorectal cancer (CRC), the third most frequent form of cancer worldwide [2], overexpression and differential expression of several ribosomal proteins have been reported [3, 4].
The contribution of the PES1-BOP1 complex, involved in ribosomal biogenesis, has been studied and showed that BOP1 is upregulated in CRC. BOP1 upregulation is associated with increased gene copy number suggesting that BOP1 overexpression may be one of the main oncogenic consequences of 8q24 amplification in CRC [5].
In order to have a more general vision of the rRNA maturation process we have analyzed the expression pattern of the genes that comprise Coute’s [6] human ribosome biogenesis dynamics model and found a significant (p < 10−3) upregulation of more than 79% of the genes associated with this model when CRC adenocarcinomas are compared to matching normal mucosa.
There are two major molecular subgroups in CRC, microsatellite stable (MSS) [7] and microsatellite unstable (MSI) adenocarcinomas that represent approximately 15% of the total incidence and associate with a better prognosis [8]. In this report we show that the upregulated transcript profile was evenly distributed among MSS and MSI subgroups except for one gene, DDX27, that differs from this pattern. Correlation of expression, to chromosomal gain showed that only 20% of the transcriptional upregulation in MSS specimens were linked to a possible gene dosage.
We identified several transcription factors (TFs) with putative binding sites overrepresented in the promoters of the ribosome biogenesis genes when compared to all genes.
In conclusion, the observed transcriptional upregulation of multiple rRNA processing genes may favour the enhanced production of fully processed rRNA that in combination with the protein components may enable the overproduction of mature ribosomal structures in CRC.
2. Materials and Methods
In a previous microarray transcript profiling study [9] we analyzed 168 colorectal tissue samples and ten normal mucosa biopsies from the resection edge (Table 1(a)). Microsatellite status determination and sample processing were carried out as described [9]. Biotin labeled cRNA was prepared from 10 μg of total RNA and hybridized to the Human Genome U133plus2.0 GeneChip (Affymetrix) containing >55.000 probe sets. The readings from the quantitative scanning were analyzed by the Affymetrix Software MAS5.0. The resulting cell files for all 178 samples were imported into ArrayAssist version 3.3 (Stratagene) and data were normalized using GC-RMA.
| Tumor specimens | 168 | Normal specimens | 10 | ||
|---|---|---|---|---|---|
| Gender | Male | 91 | Gender | Male | 8 |
| Female | 77 | Female | 2 | ||
| Age | <50 years | 12 | Age | ||
| 50–70 years | 89 | 50–70 | 6 | ||
| >70 years | 67 | >70 | 4 | ||
| Stages | MSS/MSI | ||||
| I | 1 | ||||
| II | 149 | MSS | 118 | ||
| III | 13 | MSI | 35 | ||
| IV | 5 | N.D. | 15 | ||
| Grade | 1 | 20 | |||
| 2 | 115 | ||||
| 3 | 30 | ||||
| N.D. | 3 | ||||
| Ribosome biogenesis (CRC) | Overall analysis (CRC) |
|---|---|
| T-N > 0.5 | T-N > 0.5 |
| 131/166 (78.9%) | 7237/54675 (13.2%) |
| T-N > 0.1 | T-N > 0.1 |
| 155/166 (93.4 %) | 12570/54675 (23.0%) |
| Ribosome biogenesis (bladder) | Overall analysis (bladder) | ||
|---|---|---|---|
| Ta-N > 0.5 | Ta-N > 0.1 | Ta-N > 0.5 | Ta-N > 0.1 |
| 84/123 (68.3%) | 105/123 (86.2%) | 5556/22283 (24.9%) | 7662/22283 (34.4%) |
| T2-N > 0.5 | T2-N > 0.1 | T2-N > 0.5 | T2-N > 0.1 |
| 73/123 (59.3%) | 97/123 (78.8%) | 5174/22283 (23.2%) | 7283/22283 (32.7%) |
The expression profile of the human model of ribosome biogenesis genes [6] was analysed. Those probes that did not correctly matched the NCBI data base sequences were excluded from further analysis. Median log2 values and standard deviations from CRC samples and normal matching mucosa biopsies were calculated for the set of studied genes. When two or more probes from the same gene were available a mean value was used.
The 170 genes cited in the human model of ribosome biogenesis dynamics [6] were used for chromosomal distribution analysis and transcriptome correlation map studies and 166 genes were analyzed in the microarray study due to probe restrictions.
2.1. Transcription Factor Analysis
An analysis for enrichment of transcription factor binding sites in the promoters of the 166 genes mentioned above was performed using the java-based tool Expander4.0.1 (Expression Analyzer and DisplayER). Expander utilizes the PRIMA (PRomoter Integration in Microarray Analysis) software to identify transcription factors whose binding sites are significantly overrepresented in a given set of promoters. All genes were used as background in the analysis. The promoter region analysed for each gene started 1000 bases upstream the transcription start position and ended 200 bases downstream the start position. The threshold P-value was set to .0001. The program was run with no multiple tests correction (default setting). Bonferroni correction and a threshold P-value of .001 retrieved the same transcription factors.
2.2. Correlation Study
For the 106 genes that contain at least one putative binding site for NRF1, HIF1A or ELK1, we added the number of transcription factor binding sites (TFBSs) for each of these three TFs. A regression analysis was done to study the correlation between the 106 genes overexpression and the number of TFBSs in their promoter regions.
3. Results
We have studied the transcription profile of the genes involved in ribosome biogenesis dynamics as described in Coute’s model [6] in CRC adenocarcinomas. In our study 168 CRC specimens, mostly stage II, were compared to 10 normal matching mucosas (Table 1(a)). Strikingly 78.9% of the 166 genes studied were significantly (p < 10−3) upregulated with a log2 ratio >0.5. This upregulation greatly contrasts with the overall tendency of all probes tested, as only 13.2% of the almost 55000 probes showed the same upregulation, as seen in Table 1(b).
Moreover, when we extended the study to genes upregulated with log2 ratio >0.1, up to 93.4% of the genes studied fulfilled this requirement, compared to 23% of all probes studied (Table 1(b)).
Interestingly, the gene expression upregulation profile described above was almost identical between the two major subgroups of CRC, MSS and MSI (see Supplementary Table 3 in Supplementary Material available online at doi:10-3814/2009/657042). Only DDX27, the human counterpart of yeast drs1, was significantly (p < 10−7) upregulated in MSS compared to matching normal mucosa (log2 ratio 1.2), but not in MSI specimens (p = 0.1).
In MSS specimens the observed transcript upregulation could be caused by a gene dosage effect derived from chromosomal amplification. We mapped the studied genes to a chromosomal position in a transcriptome correlation map (unpublished results), and found that only 20% could be located in areas susceptible of chromosomal gain.
We also investigated the genes chromosomal distribution. The genes involved in ribosome biogenesis dynamics were distributed along the 23 human chromosomes with no obvious clustering. The highest number of genes (15) mapped to chromosome 2. No genes were mapped to chromosome 18.
We then analysed the promoters of the studied genes for transcription factor binding sites. Four transcription factors, NRF1 (M00055[Nrf-1]), HIF1A (M00976[AHRHIF]), ELK1 (M00025[Elk-1]) and MYCN (M00055[N-Myc]), showed a significant (p < 10−6) enrichment of their putative binding sites (frequency ratio) in the promoters of the genes studied compared with all genes. Out of 166 genes, 71 had at least one TFBS for NRF1, 50 for MYCN, 45 for ELK1 and 53 for HIF1A (Table 3). Our transcript microarray analysis showed that NRF1 and ELK1 were significantly upregulated, while H1F1A was highly expressed and slightly upregulated, (Table 3). MYCN was not detected in our data set, and therefore, it was not used in the subsequent correlation study.
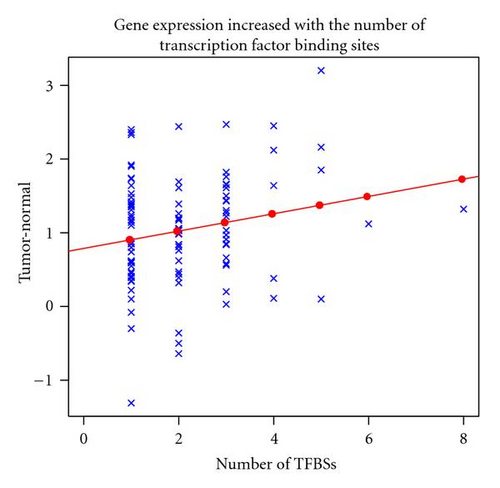
Interestingly, there was a significant linear correlation (p = 0.03) between the number of targets for NRF1, HIF1A and ELK1 in the promoters of the ribosome biogenesis genes and the level of transcript upregulation observed (Figure 1). The genes with more putative TFBSs (either several targets for the same TF or targets for more than one TF) showed a higher log2 ratio value than those with none (Figure 1 and Table 2).
| Gene | Chromosomal location | Log 2 T-N | NRF1 | ELK1 | HIF1A | MYCN* |
|---|---|---|---|---|---|---|
| EXOSC5 | 19q13.1 | 3.20 | X(1) | X(1) | X(3) | X(2) |
| RUVBL1 | 3q21 | 2.53 | — | — | — | X(1) |
| NOC4L | 12q24.33 | 2.47 | X(1) | — | X(2) | X(1) |
| BOP1 | 8q24.3 | 2.45 | X(2) | — | X(2) | X(1) |
| RRP9 | 3p21.1 | 2.44 | X(1) | X(1) | — | X(1) |
| RRS1 | 8q13.1 | 2.40 | — | X(1) | — | X(1) |
| BYSL | 6p21.1 | 2.37 | — | — | — | — |
| NOL1 | 2p25.1 | 2.36 | X(1) | — | — | X(1) |
| TBL3 | 16p13.3 | 2.33 | — | — | X(1) | — |
| WDR74 | 11q12.3 | 2.25 | — | — | — | — |
| RRP1 | 21q22.3 | 2.16 | X(4) | — | X(1) | — |
| DKC1 | Xq28 | 2.12 | X(3) | — | X(1) | — |
| DDX56 | 7p13 | 2.00 | — | — | — | — |
| EXOSC4 | 8q24.3 | 1.94 | — | — | — | — |
| PPAN | 19p13 | 1.93 | — | — | — | — |
| CGI-96 | 22q13.2-q13.31 | 1,92 | X(1) | — | — | — |
| POLR2E | 19p13.3 | 1,90 | X(1) | — | — | — |
| MRTO4 | 1p36.13 | 1,86 | — | — | — | |
| NOC2L | 1p36.33 | 1,85 | X(2) | X(1) | X(2) | — |
| SURF6 | 9q34.2 | 1,82 | — | X(2) | X(1) | X(2) |
| Set Frequency (%) | Frequency Ratio | p-value | Log 2 T-N | |
|---|---|---|---|---|
| NRF1 | 42.77 | 1.85 | <10−7 | 0.5 p < 10−5 |
| HIF1A | 31.93 | 1.76 | <10−6 | 0.2 p = .5 |
| ELK1 | 27.11 | 2.09 | <10−6 | 0.7 p < 10−5 |
| MYCN | 30.12 | 2.12 | <10−7 | u.d |
4. Discussion
The role of protein translation in the cancer process is far from well understood. In cancer cells with enhanced cell division rates and with abnormal cellular activity, the amount of proteins should increase and concomitantly the ribosome presence. However, translation and ribosome biogenesis are very well controlled processes.
Individual ribosomal proteins Sa, S8, S12, S18, S24, L13a, L18, L28, L32 and L35a have been shown to be upregulated in CRC [3] and S3, S6 L5 have also been observed upregulated at both the protein and mRNA levels at the early stages of tumorigenesis [4]. We have considered all genes involved in ribosome biogenesis in a transcript gene profile. Our results clearly showed that in CRC the majority of genes involved in ribosomal biogenesis are overexpressed at the transcriptional level.
It is tempting to argue that this upregulation could be a consequence of the abnormal growth as pointed out above. However, there are already reports indicating that some of these genes are involved in tumorigenesis. For instance, besides BOP1, WRN depletion in cancer cells inhibits tumor growth, RUVBL1 modulates transformation and apoptosis with a functional role in MYC-mediated oncogenesis, NOLA2 is associated with MYC-induced tumorigenesis, NPM1 can contribute to oncogenesis through many mechanisms, and RRP1 is related to metastasis [10–14].
Even more compelling is the fact that the regulation of rDNA transcription is critically altered in cancer. Conditions that harm metabolism, such as starvation, toxic lesions, aging, cancer or viral infections downregulate rDNA transcription [15]. However, the regulatory mechanism that would impair this downregulation is abrogated in cancer cells. The upregulation of genes that activate RNA pol I could be one of the mechanisms that help the rDNA dysregulation [15]. Our microarray analysis indicates that the transcript of RNA pol I subunits and proteins that directly modulate its activity, such as UBF, TIF1A, TIF1B and TBP are also transcriptionally upregulated (Supplementary Table 1). In addition, UBF related activating kinases casein kinase II (CKII), CDK4, cyclinD1, CDK2 and cyclin E [16] are also upregulated in CRC (Supplementary Table 1), thus allowing a possible increase in rRNA synthesis. Interestingly, a possible activation of UBTF via ERK1/2 does not seem to be favored as both kinases seem to be downregulated in colon cancer [17].
Moreover, the mechanism behind this general upregulation seems to be independent of the MSS status as only 20% of the studied genes mapped to chromosomal areas susceptible of gene amplification. When we considered the genes with highest upregulated log 2 ratio values (Table 2), only BOP1, TBL3, WDR74, EXOSC4, MRTO4 and NOC2L were located in chromosomal areas that can be subjected to gene dosage alteration. The same genes were also found highly upregulated in MSI specimens that are chromosomal stable, suggesting that the detected upregulation is accomplished by other mechanisms besides amplification. These data are consistent with published data indicating that BOP1 contributes to colorectal carcinogenesis and its overexpression is associated not only with a dosage increase of the individual gene but also with other mechanisms [5].
Apparently, not individual genes, but the whole machinery responsible for transcription of rDNA and its processing, including associated and regulatory factors, RNA polymerase I, the processome, the exosome, and several processing factors, are upregulated at the transcriptional level in CRC.
The upregulation of genes described in CRC was analysed in a similar bladder cancer gene expression profile carried out on U133A affymetrix arrays [10–12]. The results were comparable (Supplementary Table 2). Interestingly, out of the 15 most upregulated genes in the bladder profile analysis only two, DDX56 and DKC1 (Supplementary Table 2A), were also found among the 20 highest upregulated genes in the CRC study. Moreover, the TFs that were upregulated in CRC (NRF1 and ELK1) were downregulated in bladder (Supplementary Table 2B). In contrast to CRC where eight of the ten highest upregulated genes Table 2 showed putative TFBSs for the studied TFs, in the bladder study only four TFs showed TFBSs (Supplementary Table 2A).
Interestingly, ELK-1 antisense oligonucleotide is capable of suppressing hepatocellular carcinoma cells [18] and the combined expression of HIF1A and EPAS1 (HIF2A) may play an important role in tumor progression and prognosis of CRC carcinomas [19]. Interestingly, even though NMYC was not expressed in our CRC assay and thus was excluded for further analysis, the ubiquitous and well-known cancer-related MYC shares its E-box binding site with NMYC. This relation was studied in neuroblastoma cells transfected with N-myc [20]. The report revealed the upregulation of a number of genes. Most of these genes were ribosomal proteins, translational factors and genes controlling rRNA maturation. Moreover, N-myc induced rRNA content rather than protein synthesis rate. It was quite revealing that as MYCN can replace MYC in transgenic mice, they studied whether NMYC downstream targets were also induced by c-myc. Eight out of twenty targets were indeed induced, including ribosomal proteins but also NCL and NPM1 [20]. In conclusion, c-MYC could also be responsible for the activation of some genes baring an NMYC binding site.
The overall transcript upregulation concerning the genes involved in ribosome biogenesis seems to be shared for both CRC and bladder cancer.
It could be interesting to see whether other cancers also show this tendency and whether cancers with slower proliferation rate, such as prostate cancer, also share a similar pattern of expression.




