Origin and Movements of Young-of-the-Year Striped Bass in the Southern Gulf of St. Lawrence, New Brunswick
Abstract
Young-of-the-year (age-0) striped bass Morone saxatilis have been observed during late summer and fall in southern Gulf of St. Lawrence estuaries that may not support striped bass spawning. In this study, we tested the hypothesis that age-0 striped bass in the Richibucto and Kouchibouguac rivers disperse there from the Miramichi River, which is located approximately 35 km north of the Kouchibouguac River and 55 km north of the Richibucto River. Beach seining of the coastline between these three rivers during summer in 1997 and 1998 confirmed the presence of age-0 striped bass in the latter half of August but not before. Age-0 fish distributions over time were consistent with movement from the Miramichi River to the Kouchibouguac and Richibucto estuaries. Microsatellite nuclear DNA and mitochondrial DNA analyses were performed to determine the genetic relatedness among striped bass from these estuaries. Age-0 striped bass from the Shubenacadie River population in the Bay of Fundy, which is known to be genetically distinct from Gulf of St. Lawrence populations, and the Hudson River, New York, were used as out-groups in the analyses. In all comparisons, significant haplotype and allelic differences were found between Gulf of St. Lawrence samples and the Shubenacadie River and Hudson River populations. However, significant haplotype or allelic differentiation was rarely observed in comparisons among samples from rivers within the Gulf of St. Lawrence. These results suggest that some of the age-0 striped bass spawned in the Miramichi River leave their natal estuary in late summer and disperse through shallow coastal waters to other estuaries in the southern Gulf of St. Lawrence.
Introduction
The striped bass Morone saxatilis is an iteroparous, anadromous, percoid fish native to the Atlantic coast of North America between the St. John's River, Florida, and the St. Lawrence River, Quebec, as well as the Gulf of Mexico coast (Scott and Scott 1988). Most striped bass research has concentrated on populations in the center of the species' range, particularly the Chesapeake Bay and the Hudson River. Striped bass in the southern Gulf of St. Lawrence (Figure 1) have been identified as a conservation priority in Canada by both federal and community-based agencies. Recent declines in the abundance of striped bass throughout the southern Gulf of St. Lawrence (Bradford et al. 1999a, 1999b), combined with the extirpation of native populations in the St. Lawrence River (Bradford et al. 1998a), Saint John River (Rulifson and Dadswell 1995; Wirgin et al. 1995; Bradford et al. 2001), and probably the Annapolis River (Douglas et al. 2002) have led to concern for the future of this species in Canadian waters.
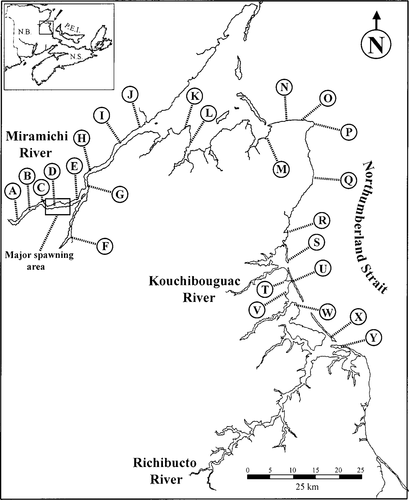
Sampling stations (letters) used in beach seining surveys in the southern Gulf of St. Lawrence, New Brunswick (N.B.), during summer 1997–1998 (inset: N.S. = Nova Scotia; P.E.I. = Prince Edward Island).
Within the southern Gulf of St. Lawrence, early research in Kouchibouguac National Park focused on a small and possibly self-sustaining population of striped bass (Melvin 1979; Hogans and Melvin 1984). The Kouchibouguac River was thought to be a spawning site for striped bass based on observations of putative spawning behavior (Hogans and Melvin 1984) and on the presence of adult fish in the estuary during spawning season (Melvin 1979). There are also records of young-of-the-year (age-0) striped bass being captured during the fall in the Kouchibouguac River (Hogans and Melvin 1984; Robinson et al. 1998). Similarly, adults were known to be present in winter and spring in the nearby Richibucto River, and age-0 fish had been observed in the Richibucto River estuary in fall (S. Courtenay, unpublished data).
Age-0 striped bass are considered to be nonmigratory throughout the species' range and are believed to be restricted to their natal rivers and estuaries until they reach 2–4 years of age, depending on their gender and stock of origin (Merriman 1941; Klauda et al. 1980; Kohlenstein 1981; Rulifson and Dadswell 1995; Secor and Piccoli 1996; Wainright et al. 1996). Additionally, it has been suggested that striped bass stocks at the southern (Dudley et al. 1977) and northern (Magnin and Beaulieu 1967) limits of the species' range are nonmigratory at all ages and are restricted throughout the life cycle to natal estuaries and proximal coastal waters. Thus, the presence of age-0 striped bass in the Kouchibouguac and Richibucto rivers implied that spawning had occurred there. The presence of documented spawning striped bass populations in these rivers would impact local fishery management strategies.
Ichthyoplankton and beach seining surveys in the Kouchibouguac River in spring and early summer of 1996 found no evidence of striped bass spawning, although age-0 striped bass were present in the estuary in late summer (Robinson et al. 1998). Robinson et al. (1998) suggested that these fish originated from a nearby river, such as the Miramichi to the north or the Richibucto to the south. Intensive ichthyoplankton surveys (a total of 50) in the Kouchibouguac and Richibucto rivers in 1997 and 1998 found no evidence of striped bass spawning in either river, although age-0 fish appeared in the lower estuaries of both rivers during late summer (Robinson et al. 2001). The Kouchibouguac and Richibucto river surveys were conducted twice weekly during May and June and biweekly thereafter through August of 1997 and 1998. The ichthyoplankton community was sampled both upstream and downstream of the salt front, where striped bass spawning was expected to occur (Robinson et al. 2001). Although no striped bass eggs or larvae were captured during these surveys, the anadromous fish community (such as rainbow smelt Osmerus mordax, alewives Alosa pseudoharengus [also known as gaspereau], and blueblack herring A. aestivalis) was well represented, indicating that the gear was effective for the collection of pelagic spawners such as striped bass. Age-0 striped bass were found in these estuaries throughout the fall (S. Courtenay, unpublished data) and have been observed overwintering with older conspecifics near the heads of these estuaries (R.G. Bradford, Fisheries and Oceans Canada, Dartmouth, Nova Scotia).
The Miramichi River estuary is the only confirmed (i.e., eggs and larvae documented and described) striped bass spawning site in the southern Gulf of St. Lawrence. Studies in this river have demonstrated that larval and juvenile striped bass undergo a series of predictable habitat shifts during their first few months of life (Robichaud-LeBlanc et al. 1996, 1998). Spawning takes place during the early spring immediately upstream of the saltwater intrusion (Figure 1). The larvae occupy pelagic riverine habitat for approximately 1 month until yolk sac absorption is complete, after which the juveniles move onshore and utilize littoral habitats. As the summer progresses, age-0 striped bass begin to move into progressively more saline littoral waters until they occupy the more saline reaches of their natal estuary (Robichaud-LeBlanc et al. 1998).
The objective of this study was to test the hypothesis that age-0 striped bass present in the Kouchibouguac and Richibucto estuaries in late summer disperse there from the Miramichi River. The timing of this movement was examined through coastal beach seining surveys in the littoral zone between the Miramichi and Richibucto estuaries; the surveys were executed before and during the months that age-0 striped bass were observed moving into the Kouchibouguac estuary in 1996 (Robinson et al. 1998). The genetic identity of age-0 striped bass entering the Kouchibouguac and Richibucto estuaries was compared to that of age-0 fish in the Miramichi River by use of microsatellite nuclear DNA (nDNA) and mitochondrial DNA (mtDNA) markers. Previously, analysis of mtDNA detected significant haplotype differences between striped bass populations in the Gulf of St. Lawrence (Miramichi and Tabusintac rivers) and the Shubenacadie River in the Bay of Fundy, but found an absence of population structure among rivers within the Gulf of St. Lawrence (Wirgin et al. 1993, 1995). Similarly, the Shubenacadie River population was genetically distinct from U.S. populations. The present study represents the first application of microsatellite nDNA technology to Canadian striped bass.
Study Area
The Kouchibouguac River estuary (Figure 1) is small, draining a catchment basin of approximately 228 km2 (Ambler 1975; J. Kerekes, Canadian Wildlife Service, unpublished report), and it has a mean annual freshwater discharge rate of approximately 3.74 m3/s (Beach 1988). Depending on the amount of surface runoff present, tidal effects in the system can extend as far as 15 km upstream to a derelict hydroelectric dam located in Kouchibouguac Village (Kerekes, unpublished).
The Richibucto River estuary (Figure 1) is located approximately 20 km south of the Kouchibouguac River estuary. It is a small coastal watershed with a mean annual freshwater discharge rate of 26.0 m3/s and a maximum discharge rate of approximately 91.5 m3/s (St. Hilaire et al. 1997). The Kouchibouguac and Richibucto rivers both drain into shallow coastal lagoons that are separated from the Northumberland Strait by a 25-km-long procession of barrier sand-dune islands, which buffer the estuaries from the more dynamic conditions of the strait. The two estuaries have traditionally supported large annual catches of striped bass as bycatch of the smelt and gaspereau fisheries (LeBlanc and Chaput 1991) and continue to offer important overwintering habitat for striped bass (Bradford et al. 1998b).
Methods
Beach seining
Age-0 striped bass were collected with a 25-m × 1.5-m, bag-style seine constructed with 6-mm mesh. The 3.4-m3 purse of the net was fitted with a 0.9-mm nylon mesh liner to retain fish eggs and larvae. The beach seine was deployed by securing one end to the shore while the other end was manually towed perpendicular from shore for a distance of approximately 15–20 m. The seine was then brought into shore in a quarter-circle sweep, usually against the current. An area of approximately 240 m2 and depth of 1.5 m was sampled with this method. At each sampling site, surface salinity (SS) was recorded approximately 2 m from shore by use of a hand-held refractometer (#A366 ATC, Ben Meadows Co., No. 221192, Atlanta, Georgia). All captured fish were identified to species, and any striped bass with total lengths (TL) less than 100 mm were identified as age-0 fish (Robichaud-LeBlanc et al. 1998).
Timing of beach seining
Two types of beach seining surveys, coastal and riverine, were used in this study. These surveys were designed to maximize sampling effort in areas where age-0 striped bass would likely be distributed if spawning had taken place. Based on previous studies in the Miramichi estuary (Robichaud-LeBlanc et al. 1996, 1998), we assumed that if striped bass spawning had occurred in the Kouchibouguac and Richibucto rivers, the age-0 fish would be distributed in littoral habitat with SS of 0–5‰ in June, 0–15‰ in July, and 0–25‰ in August.
The coastal beach seine surveys sampled the littoral zone from the known striped bass spawning grounds in the Miramichi River to the Richibucto estuary (Figure 1). In 1997, four coastal beach seine surveys were carried out from August 12 to 27. In 1998, 11 coastal seining surveys were conducted from July 8 to August 25.
The riverine surveys sampled the Kouchibouguac and Richibucto rivers (Figure 2A, B) a total of 13 times in 1997 and 25 times in 1998. During the early summer, when we expected any locally spawned age-0 striped bass to be present in the upper reaches of the river estuaries, the beach seining surveys concentrated on sampling the upper riverine littoral habitat. During the late summer, when we expected age-0 striped bass to begin a shift to more saline waters, the beach seining effort was concentrated further downriver in both the Kouchibouguac and Richibucto systems.
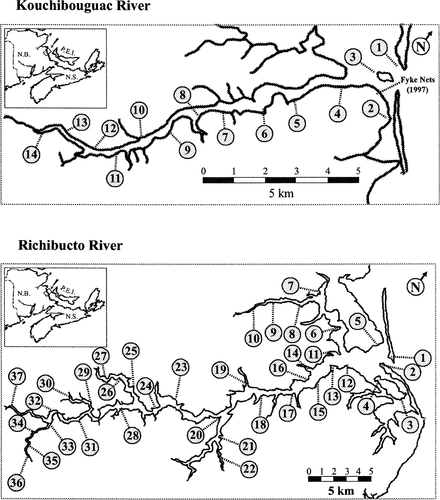
Beach seining sites surveyed in the Kouchibouguac and Richibucto rivers, 1997–1998. The fyke-net array, sampled in the Kouchibouguac River in 1997, is also indicated
In late August of 1997, juvenile fish were also sampled in the Kouchibouguac River with a linear array of four fyke nets positioned near the river mouth (Figure 2). The fyke nets, specially constructed to capture and retain small fishes, consisted of 1-cm stretch mesh and 6-m leaders. The nets were set in a slightly staggered line perpendicular from the shore near the mouth of the river, and were usually fished daily. These nets were used to sample offshore areas in Kouchibouguac Lagoon that were inaccessible to beach seining.
Sample collections for genetic analyses
A subset of age-0 striped bass collected in 1997 and 1998 from within-river beach seine surveys in the northwest Miramichi, Kouchibouguac, and Richibucto rivers were retained for DNA analyses. The Kouchibouguac River was difficult to seine in 1998 due to macrophytic algal blooms in the shallow littoral zone, and no age-0 striped bass were captured. Shubenacadie River and Hudson River collections were included in the analyses as positive controls for genetic differentiation. Shubenacadie River fish were captured from August 17 to September 3, 1997–1998, immediately downstream of the Highway 102 crossing of the Stewiake River tributary, approximately 2 km upstream of its confluence with the Shubenacadie River, and within 4 km of known striped bass spawning grounds further upstream (Rulifson and Tull 1999). Hudson River specimens were a subset of those described in Wirgin et al. (1993) and represented a combination of spawning adults and age-0 fish collected at or near the known Hudson River spawning grounds.
At least 20 fish were collected from each river during each year (except Kouchibouguac River in 1998), for a total of at least 40 individuals per river system. All Miramichi River fish (excluding two individuals) were caught on known striped bass spawning grounds (Robichaud-LeBlanc et al. 1996) of the northwest Miramichi River (stations A, B, and D; Figure 1) between July 8 and August 12. Kouchibouguac River samples were collected between August 18 and 26 by beach seining at station 2 and by fyke nets (Figure 2). Richibucto River specimens were caught between August 18 and 28 at stations 4, 5, 6, 7, 14, 19, 20, 24, and 25 (Figure 2). All fish were immediately preserved in 95% ethanol.
DNA isolations
Approximately 50 mg of fin tissue was used to isolate DNA (except for the Hudson River sample, from which frozen livers were used) with the standard phenol–chloroform extractions and alcohol precipitations (Sambrook et al. 1989). The DNA pellets were air-dried and rehydrated with double-distilled H2O (ddH2O). After hydration, the quality of each DNA sample was evaluated by ultraviolet spectrophotometry at 260 and 280 nm, and by electrophoretic separation of DNA aliquots in 0.8% agarose gels stained with ethidium bromide.
MtDNA analysis
Total DNAs were amplified by polymerase chain reactions (PCR) with primers F2 5′-CCTTTGTGTCACAGA AT-3′ and R2 5′-TCAACTCTCAGGATAACG-3′ (Invitrogen, Carlsbad, California) in PTC-100 thermal cyclers (MJ Research, Inc., Watertown, Massachusetts). The primers amplify a portion of the striped bass mtDNA control region that contains multiple copies of 120-bp tandem repeated sequences. Reactions were conducted in 25-μL volumes containing 1 μL of DNA (approximately 10–50 ng), 0.05 μL (1.25 units) of KlenTaq1 enzyme (AB Peptides, Inc., St. Louis, Missouri), 2.5 μL of KlenTaq1 reaction buffer, 1 μL of each primer (30-mM stock), 50 μM of each dNTP (Promega Corp., Madison, Wisconsin), and ddH2O to volume. Cycling parameters were as follows: initial denaturation at 95°C for 5 min, followed by 25 cycles at 94°C for 30 s, 54°C for 30 s, and 72°C for 60 s, and a final extension at 72°C for 7 min.
Polymerase chain reaction products were electrophoretically separated in 15-cm2 agarose gels in 1× tris–borate–EDTA buffer and run at 80 V for 4–6 h. Gels were stained in ethidium bromide and scanned with a ChemiImager 4400 Low Light Imaging System and enhanced with Graphic Converter version 3.4.1 (Alpha Innotech Corp., San Leandro, California). All gels contained a 1-kilobase (kb) molecular size marker (Invitrogen) and mtDNA samples representative of the complete suite of previously characterized striped bass mtDNA length-variant haplotypes (Wirgin et al. 1993).
Microsatellite nDNA analysis
The four microsatellites (SB91, SB108, SB113, and SB117 ) used in this study were chosen from a battery of loci developed for striped bass (Roy et al. 2000). Loci were selected based on the number of alleles observed in a subset of Canadian Atlantic striped bass, success rate of PCR amplification, and ease of locus visualization with an automated genetic analyzer.
The PCRs were conducted in 12.5-μL total volumes that contained 50 ng of total DNA, 0.5 μL of each primer (1 μM stock), 1.25 μL of 10× reaction buffer (+Mg) (Roche Applied Science, Indianapolis, Indiana), 0.1 μL of each deoxynucleotide triphosphate (dNTP) (250-μM stock of each) (Promega Corp.), 0.1 μL of Taq DNA polymerase (0.5 units) (Roche Applied Science), and ddH2O to volume. Primers at SB113 and SB91 were those listed in Roy et al. (2000). Primers were 5′-AGTCATGCAAACACCTGA-3′ and 5′-TGGAAGACTCCCTCTGTC-3′ at SB117, and 5′-ACTCTCGTATCGAACCAT-3′ and 5′-CTGGTCAAGCCTTTACTG-3′ at SB108. One primer at each locus was fluorescently labeled (D2-D4/PA) (Research Genetics, Huntsville, Alabama), and the second primer was unlabeled. The PCR amplifications were performed in PTC-100 thermal cyclers. Cycling parameters were as follows: denaturation at 95°C for 5 min, followed by 50 cycles of denaturation at 94°C for 60 s, annealing at 56°C (SB91, SB113, and SB117) or 54°C (SB108) for 60 s, extension at 72°C for 60 s, and a final extension at 72°C for 7 min.
Depending upon the success of amplifications (determined empirically by testing a subset of amplicons in agarose gel electrophoresis), PCR reactions were diluted up to 1:3 with sample loading solution (Beckman Coulter, Fullerton, California), and 0.5–2 μL of diluted PCR reactions were loaded onto 96 well plates along with 0.5 μL of CEQ DNA Size Standard 400 (Beckman Coulter) and 40 μL of sample loading solution. Reactions were overlaid with mineral oil and loaded into a CEQ 2000 XL automated sequencer (Beckman Coulter) and run with the FRAG 1 program.
Statistical analysis
Tests for genetic homogeneity of mtDNA haplotype frequencies were conducted in the MONTE module of the REAP 3.1 software package (McElroy et al. 1992). MONTE employs a Monte Carlo X2-based method to test for genetic homogeneity between pairs of populations (Roff and Bentzen 1989). All Monte Carlo simulations were run with 10,000 iterations and were tested against a critical value of 0.05.
Deviations from Hardy–Weinberg proportions at microsatellite loci were examined by use of GENEPOP 3.1 (Raymond and Rousset 1995). Deviations from Hardy–Weinberg equilibrium proportions can indicate nonrandom mating or small populations (Weir 1996), selection, or population mixing (Rousset and Raymond 1995). Allelic frequency data were examined for heterozygote deficit with the score (U) test developed for multiple-sample analysis by Rousset and Raymond (1995). Significant P-values produced from this test (α = 0.05) are indicative of nonrandom union of gametes. Linkage disequilibrium was tested by use of the linkage disequilibrium option in GENEPOP 3.1 (Raymond and Rousset 1995). Tests of population differentiation were performed in Arlequin 1.1 Genetic Data Analysis Software (Schneider et al. 1997) using pairwise fixation index (FST) comparisons, and in GENEPOP 3.1 (Raymond and Rousset 1995) using genic-based (Fisher's exact test) and genotypic-based (log-likelihood-based exact test) pairwise comparisons. Population assignment tests were conducted in the WhichRun software package (available at http://www-bml.ucdavis.edu/whichrun.htm). Bonferroni adjustments were applied to the P-values generated from all component tests (Rice 1989) to guard against type I errors.
Results
Age-0 Striped Bass Distribution
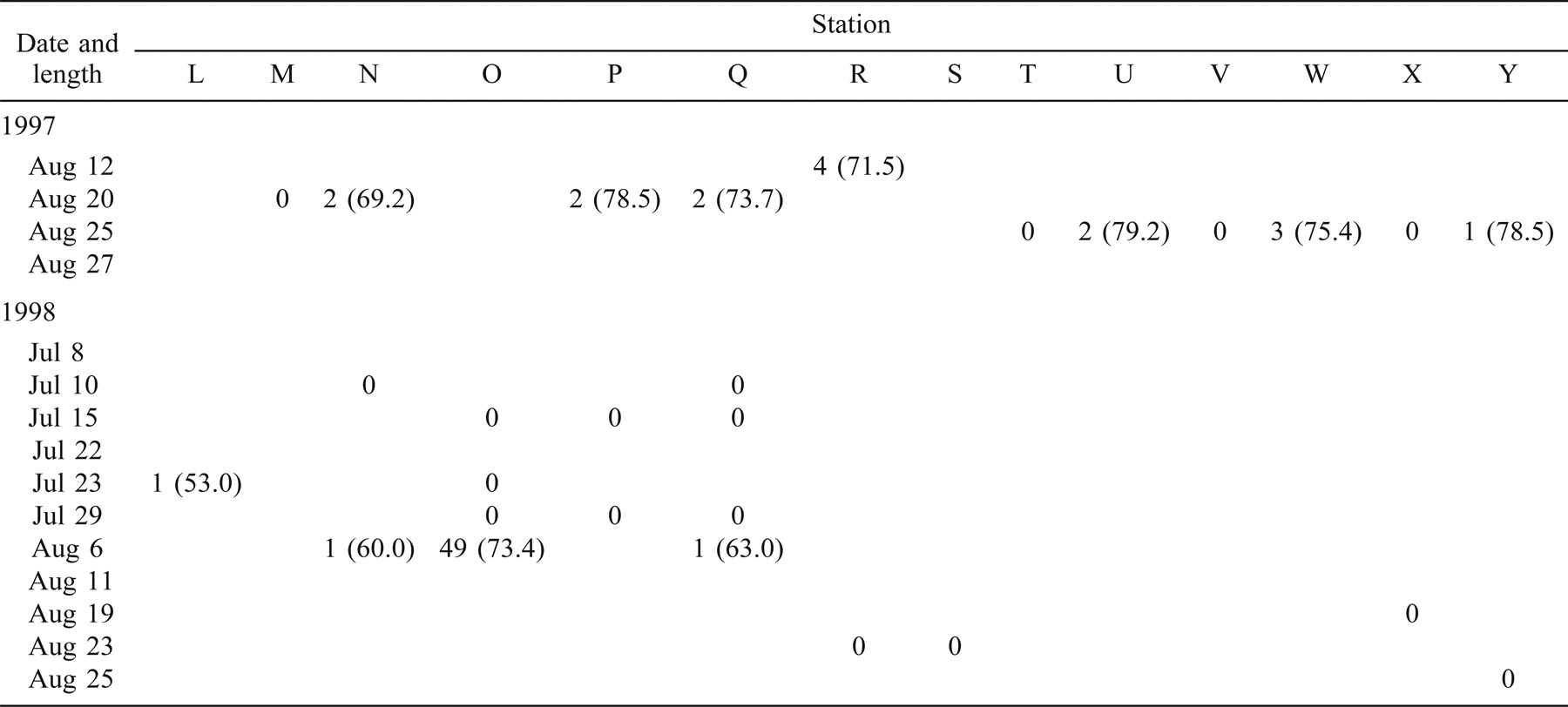
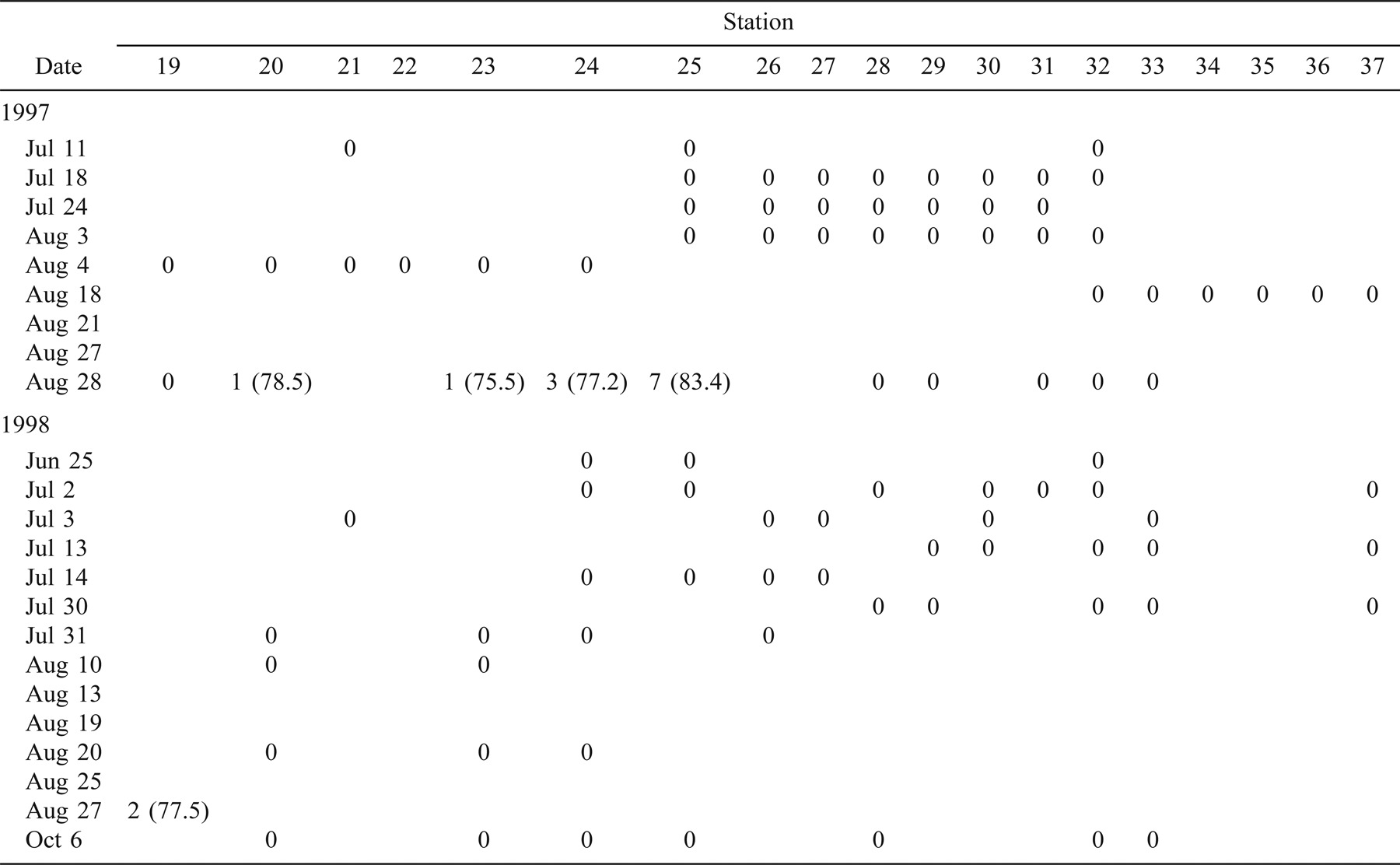
In 1997, 330 age-0 striped bass were captured during beach seining between the spawning grounds in the Northwest Miramichi River and the Richibucto estuary (Table 1). Of these, 314 were seined from the Miramichi River on August 12 in waters with SS values ranging from 0‰ to 7‰. The mean lengths of these fish at individual sampling stations ranged from 31.2 to 59.8 mm TL (Table 1). Age-0 striped bass were subsequently captured in the Kouchibouguac (Table 2) and Richibucto estuaries (Table 3). On August 19, 62 age-0 striped bass were captured within the Kouchibouguac estuary near the mouth of the river (Table 2). These fish were captured at station 2 (SS = 27‰; N = 3, mean TL = 71.5 mm) and station 4 (SS = 25‰; N = 59, mean TL = 68.5 mm). Fyke nets positioned near the mouth of the Kouchibouguac River (SS > 25‰) captured age-0 striped bass on August 20 (N = 19), August 21 (N = 37), August 22 (N = 2), and August 27 (N = 2) (Table 2). Age-0 striped bass were subsequently captured in the Richibucto estuary on August 21 (SS = 8–25‰; N = 15), August 27 (SS = 5‰; N = 2), and August 28 (SS = 11–21‰; N = 12) (Table 3). Continued beach seine sampling in coastal locations on August 25, 1997 (Table 1), captured age-0 striped bass at station N (SS = 23‰; N = 2, mean TL = 69.2 mm), station P (SS = 27‰; N = 2, mean TL = 78.5 mm), and station Q (SS = 27‰; N = 2, mean TL = 73.7 mm) (Table 1). By the end of the sampling period in late August of 1997, age-0 striped bass were collected in the Richibucto River as far upriver as station 25, at the mouth of the Molus River (SS = 11‰; N = 7, mean TL = 83.4 mm) (Table 3).
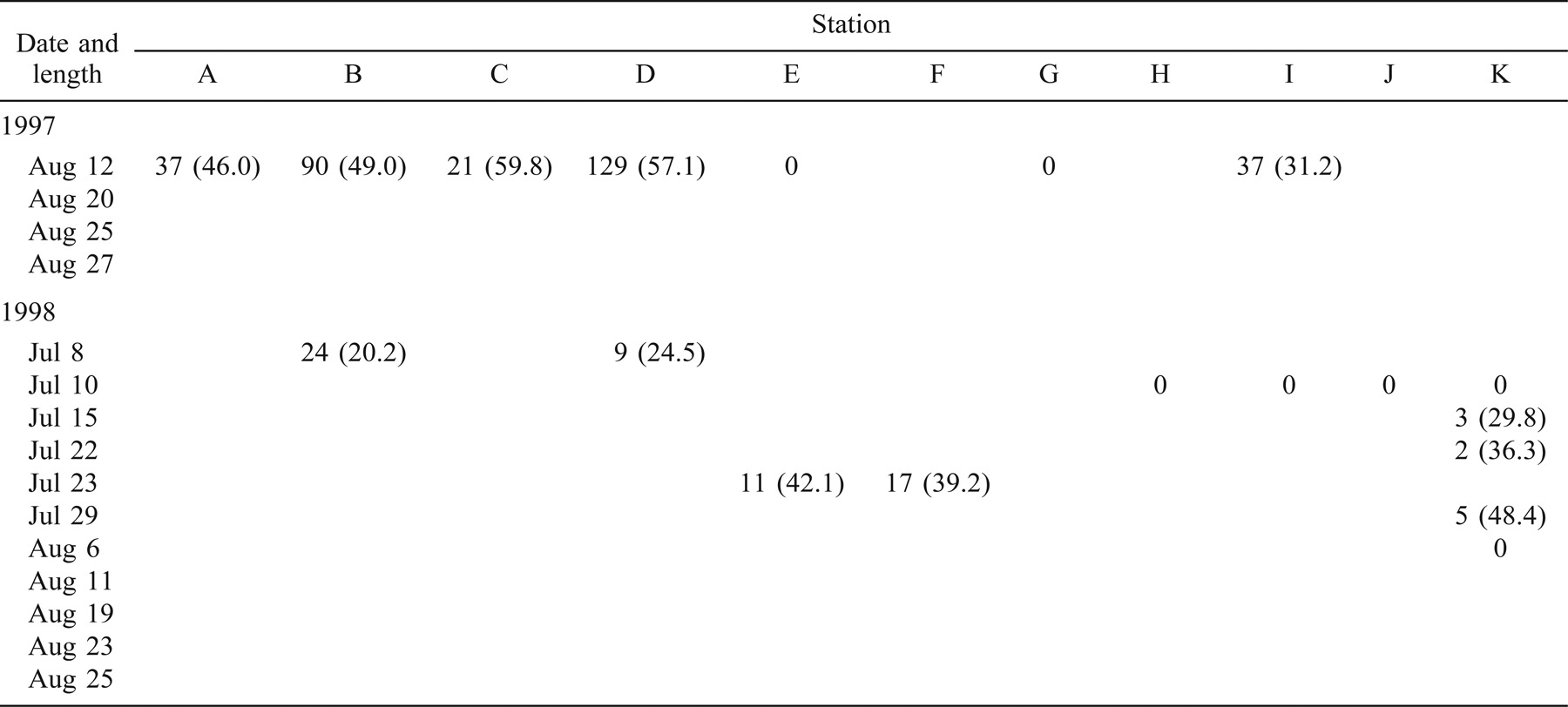
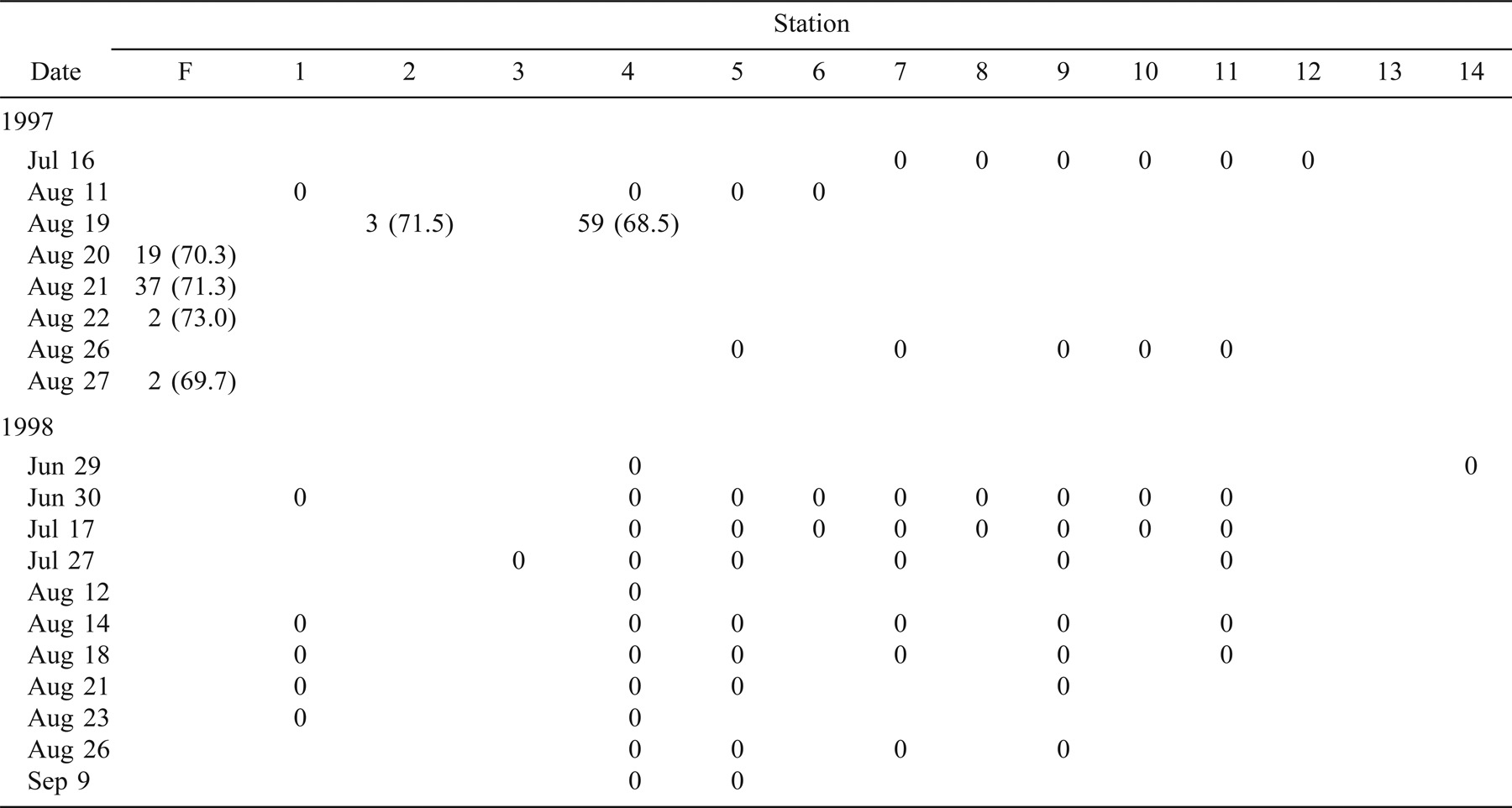
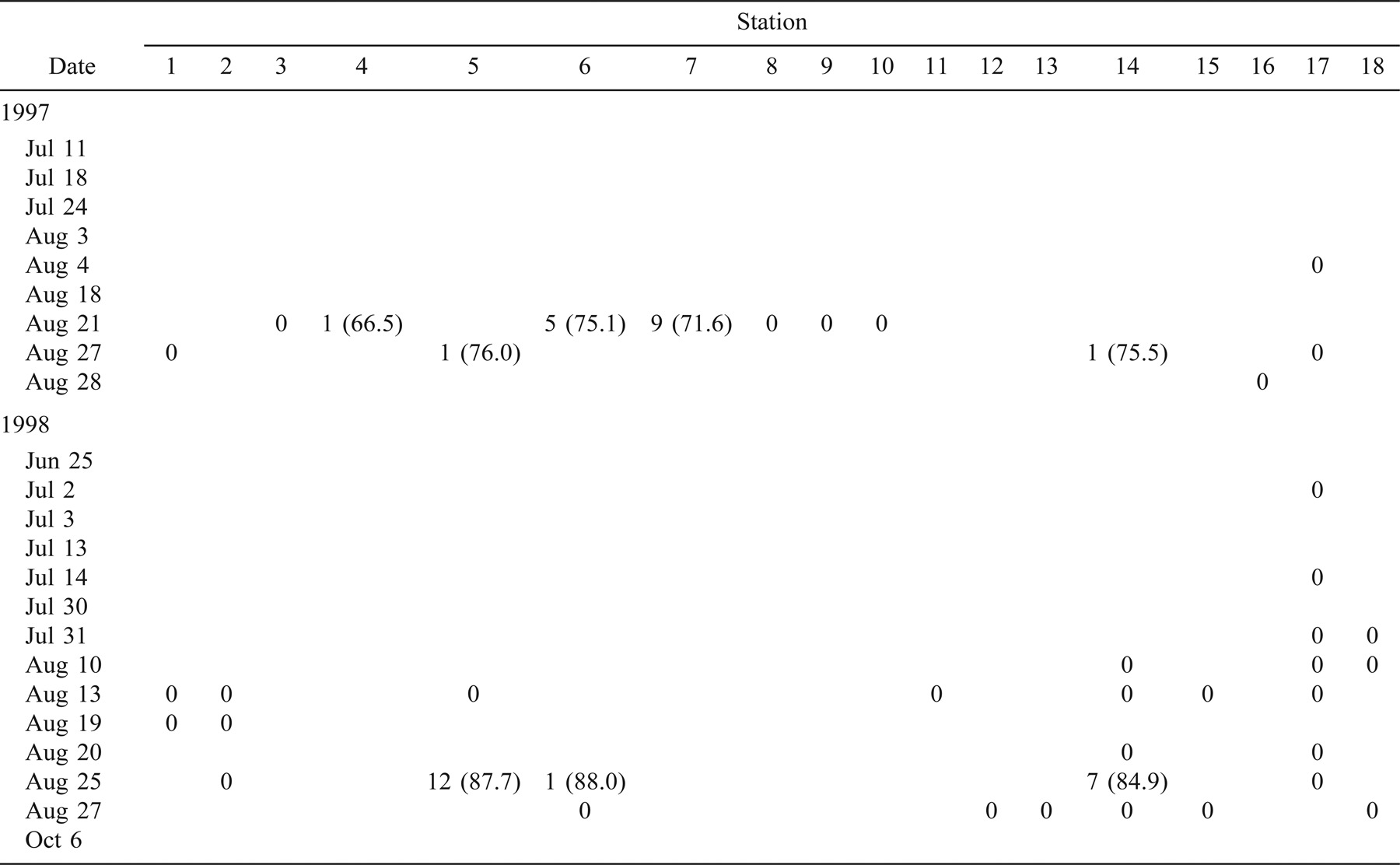
In 1998, 123 age-0 fish were captured during the coastal beach seining surveys (Table 1). Of these, 61 were captured from the northwest and southwest Miramichi rivers on July 8 and 23 (SS = 0–3‰). These fish ranged in length from 16.4 to 48.5 mm TL. Age-0 striped bass were first captured outside of the Miramichi estuary on August 11, when 49 individuals (mean TL = 73.4 mm) were collected at station O (SS = 26‰). Age-0 striped bass were also sampled on this day at station N (SS = 26‰; N = 1, TL = 60.0 mm) and at station Q (SS = 28‰; N = 1, TL = 63.0 mm) (Table 1). The coastal area where these three sampling stations are located was also seined on July 15, 22, and 29, and on August 6 (Table 1), but no age-0 striped bass were captured. No age-0 striped bass were captured in the Kouchibouguac estuary during the 11 beach seining surveys conducted between June 29 and September 9, 1998 (Table 2). A total of 22 age-0 striped bass were captured in the Richibucto estuary between August 25 and 27, 1998 (Table 3), at SS ranging from 22‰ to 26‰. The smallest of these, measuring 71.0 mm TL, was captured at station 5 near the mouth of the northwest arm of the Richibucto River (Table 3). During late August of 1998, age-0 striped bass were present in the Richibucto River at least as far upriver as station 19, near the mouth of Mill Creek (Table 3).
Mitochondrial DNA Analysis
Frequencies of mtDNA length haplotypes were similar in fish collected from the Miramichi, Richibucto, and Kouchibouguac rivers (Table 4). No significant pairwise differences in haplotype frequencies were observed between samples collected from these three rivers (X2 = 0.24–0.32, P = 0.734–0.879). The Miramichi, Richibucto, and Kouchibouguac river samples all differed significantly from the Shubenacadie River collections (X2 = 21.48–24.42, P < 0.0001). Samples analyzed from the Shubenacadie River were characterized by a high frequency (55%) of length haplotype A (Table 4), which was relatively rare in all three Gulf of St. Lawrence populations (<8%). Within-river mtDNA length haplotype frequencies determined in this study did not differ significantly from those measured in previous investigations in the Miramichi and Shubenacadie rivers (Table 4).
Microsatellite Analysis
The observed number of alleles across all populations at each microsatellite locus (Table 5) ranged from 10 (SB91) to 33 (SB113) (mean = 19 alleles). The mean number of alleles per locus within the individual river collections (over all samples) ranged from 4.4 to 16. Observed heterozygosity measurements by locus for each river sample ranged from 0.022 for SB91 in the Kouchibouguac River sample to 0.979 for SB113 in the Hudson River sample. Observed heterozygosities averaged across all four loci ranged from 0.430 (Miramichi River sample) to 0.684 (Hudson River sample). Based on observed and expected heterozygosities, levels of genetic diversity were higher for the Shubenacadie River and the Hudson River than for the southern Gulf of St. Lawrence samples. In individual locus × population tests of Hardy–Weinberg equilibrium, one significant departure was observed at locus SB117 in the Shubenacadie River sample. Tests of genotypic disequilibrium found no evidence of linkage between any of the microsatellite loci in any of the populations sampled.
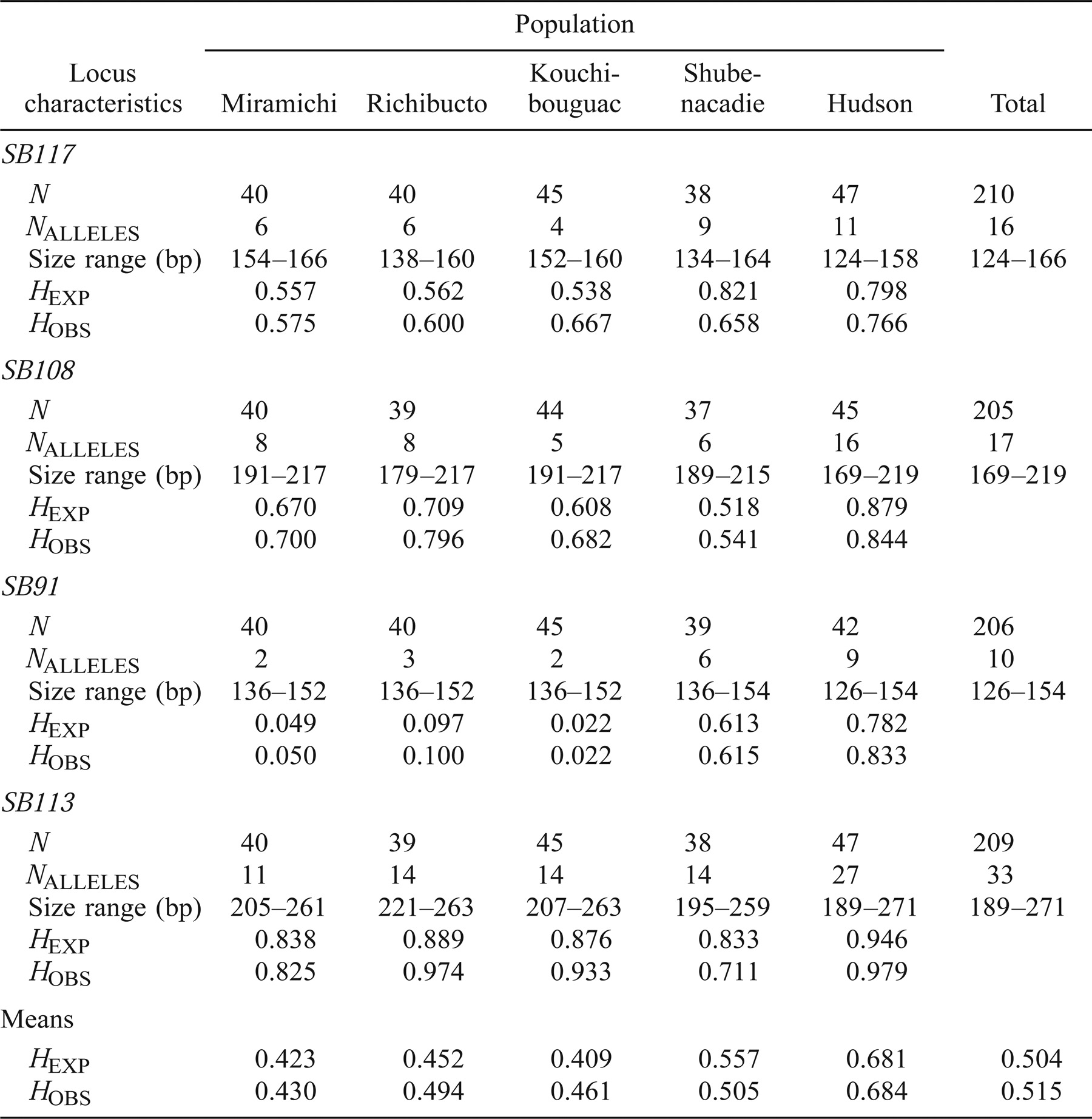
Because population samples were collected over a 2-year period in the Shubenacadie, Miramichi, and Richibucto rivers, it was necessary to test for temporal variation in these samples before they were combined over the 2-year period for each river. Within each river, neither genic nor genotypic differentiation tests across all loci detected any significant differences between the allelic or genotypic composition of river collections between the two sampling years.
Assignment tests between individuals from the Kouchibouguac, Richibucto, and Miramichi rivers failed to assign individuals to the correct river with high probability, corresponding to putative homogeneity among the Gulf of St. Lawrence samples. Assignment was low for these rivers, with correct assignment values of 32.5% (Richibucto), 42.5% (Miramichi), and 62.5% (Kouchibouguac). In contrast, assignment values were much higher for the remaining rivers, with 87.2% of Shubenacadie River and 91.8% of Hudson River individuals assigning correctly.
Pairwise FST comparisons detected consistent, highly significant genetic differences (P < 0.005) between the southern Gulf of St. Lawrence striped bass collections and the Shubenacadie River and Hudson River samples. The FST values for these pairwise tests ranged from 0.1144 (Richibucto versus Shubenacadie) to 0.2075 (Kouchibouguac versus Hudson). No significant differences were detected between samples from the three southern Gulf of St. Lawrence rivers, with FST values for these comparisons ranging from 0.0022 (Kouchibouguac versus Richibucto) to 0.0111 (Richibucto versus Miramichi).
We also used the genic heterogeneity test in GENEPOP to test for differentiation among striped bass collections, as a robust alternative to the FST comparisons. Genic tests revealed highly significant differentiation at all four loci when all collections were considered. Testing exclusively among the three Gulf of St. Lawrence samples revealed heterogeneity among collections at one locus, SB113, resulting from significant differentiation between the Kouchibouguac River and Richibucto River samples.
The extent of genetic homogeneity among Gulf of St. Lawrence samples was also examined through pooling the Gulf of St. Lawrence samples and retesting for Hardy–Weinberg equilibrium conditions and linkage disequilibrium. In this case, all loci were unlinked, but a single significant departure from Hardy–Weinberg equilibrium was detected at SB117.
Discussion
The results of this study are most consistent with the idea that some age-0 striped bass originating in the Miramichi River move into coastal waters of the Gulf of St. Lawrence at least as far south as the Richibucto estuary. This is supported by the temporal distribution of age-0 striped bass that were seined in 1998 and by the genetic data from two independently inherited genomes. Highly significant differences in nDNA microsatellite allelic frequencies at all four loci and in mtDNA haplotype frequencies were observed between all Gulf of St. Lawrence collections and those from the Shubenacadie River and Hudson River, and between the Shubenacadie River and Hudson River samples. In contrast, significant genetic differences were absent in the mitochondrial genome, and only one microsatellite locus revealed nDNA heterogeneity among collections of age-0 striped bass from the Miramichi, Richibucto, and Kouchibouguac estuaries. This suggests that most age-0 striped bass in the Richibucto and Kouchibouguac rivers originate from the Miramichi River.
The data collected during the 1997 coastal beach seining surveys cannot be used to determine the direction of the putative coastal movements because no coastal baseline data were collected prior to the appearance of the age-0 striped bass within the Richibucto and Kouchibouguac estuaries. In 1997, nonestuarine coastal areas in the Northumberland Strait, such as Point Escuminac (station P, Figure 1) and Pointe Sapin (station Q, Figure 1), were not sampled until the presence of age-0 striped bass had already been detected in the Kouchibouguac and Richibucto rivers.
Data on the presence of age-0 striped bass were collected in the nonestuarine areas during the coastal beach seining surveys in 1998. Repeated beach seining surveys beginning on July 15 failed to capture age-0 striped bass in the Point Escuminac–Pointe Sapin region until August 11. In 1998, no age-0 striped bass were captured in the Richibucto estuary until August 25, despite an intensive beach seining effort. This pattern of appearances is consistent with a southerly movement of age-0 fish from the Miramichi estuary, and supports the hypothesis that age-0 striped bass collected in the Richibucto and Kouchibouguac estuaries exited from the Miramichi River and moved through coastal waters to these other systems. This is supported by the temporal appearance of age-0 striped bass within the Richibucto estuary in 1997 and 1998 and within the Kouchibouguac estuary in 1997. Of course, it is possible that our sampling effort within these two estuaries may have been insufficient to detect age-0 fish prior to these dates.
The results from this study are congruent with those from previous investigations, based on mtDNA length-variant haplotypes, of the genetic relatedness of striped bass from rivers in the southern Gulf of St. Lawrence and Bay of Fundy and the U.S. coastal migratory stock. Highly significant differences in mtDNA length-variant haplotype frequencies were observed between striped bass from the Miramichi River and the Tabusintac River in the Gulf of St. Lawrence compared to fish from the Shubenacadie River in the Bay of Fundy (Wirgin et al. 1993) and between Shubenacadie River fish and U.S. coastal migrants (Wirgin et al. 1993, 1995). Our current mtDNA results lend further support to an absence of mixing between striped bass populations from the Bay of Fundy and the southern Gulf of St. Lawrence. In contrast, mtDNA length haplotype frequencies were very similar among collections of age-0 striped bass from three rivers in the southern Gulf of St. Lawrence. Our study strengthens the conclusions of Wirgin et al. (1993) in that our mtDNA analysis strictly focused on age-0 fish, as opposed to the yearlings that were analyzed in the previous study. Additionally, frequencies of mtDNA length haplotypes in age-0 striped bass collected from the Miramichi River in 1993 (I. Wirgin and S. Courtenay, unpublished data) were very similar to those in yearlings collected in 1990 (Wirgin et al. 1993) and those in age-0 fish collected in the current study. Similarly, mtDNA length haplotypes in the age-0 striped bass we collected from the Shubenacadie River in 1997 and 1998 are very similar to those in age-0 striped bass collected in the Shubenacadie River in 1991 and 1992 (Wirgin et al. 1993, 1995). These results point to the temporal stability of mtDNA length haplotypes in striped bass from these Canadian rivers.
Our microsatellite results are completely congruent with the mtDNA data in defining the genetic relatedness among striped bass collections from the Gulf of St. Lawrence, the Shubenacadie River, and the Hudson River. All population × locus comparisons among these three collections revealed highly significant allelic differences. Thus, based on data from two genomes, there are at least two genetically distinct stocks of striped bass in Canadian waters, Gulf of St. Lawrence and Shubenacadie River, both of which are genetically dissimilar from the U.S. coastal migratory stock.
Our microsatellite results are generally consistent with mtDNA results in failing to detect significant genetic divisions among rivers in the southern Gulf of St. Lawrence. The FST comparisons among the three individual Gulf of St. Lawrence collections failed in all cases to detect significant heterogeneity. Additionally, an absence of linkage disequilibrium and the conformance to Hardy–Weinberg expectations (with one exception) in the pooled Gulf of St. Lawrence collection support the existence of a single striped bass population in the southern Gulf of St. Lawrence.
The extent of genetic differentiation observed among the southern Gulf of St. Lawrence, Shubenacadie River, and Hudson River samples suggests that the microsatellite analysis was sensitive to genetic divergence between some populations that had only recently been reproductively isolated, such as those in this study. These populations have likely been reproductively isolated from one another only since recolonization following the recession of the Wisconsin Glaciation, approximately 10,000 years ago (Schmidt 1986). However, it should be recognized that an absence of genetic divergence among river collections within the Gulf of St. Lawrence, even in two independently transmitted genomes, does not necessarily indicate that they currently form one homogeneous reproductive unit. It is possible that reproductive isolation does exist among rivers in the Gulf of St. Lawrence but has not yet been manifested as significant haplotype or allelic discontinuities. Also, the limited number of loci that we investigated may have been insufficient to detect significant differences among populations that only recently diverged.
Although the weight of evidence supports the genetic homogeneity of the Gulf of St. Lawrence collections, the more stringent genic test did reveal heterogeneity at one locus, SB113, between the Kouchibouguac River and Richibucto River collections, although not between either of these two rivers and the Miramichi River. One explanation is that age-0 fish that disperse from the Miramichi River to other rivers only represent a fraction of genotypes within the Miramichi spawning population. This would be consistent with Secor's (1999) contingent hypothesis, in which metapopulations such as that of the Miramichi River are composed of several smaller contingents that may exhibit differing life history characteristics. However, the observed heterozygosity and number of alleles in the Kouchibouguac and Richibucto samples were not less than those observed in the Miramichi collection. Similarly, when population density in the Miramichi River is high, spillover of spawning fish may occur and spawning may take place in southern Gulf of St. Lawrence rivers other than the Miramichi. This scenario is frequently observed in wide-ranging species such as striped bass, and may in fact account for the re-establishment of striped bass spawning in the extirpated or nearly extirpated Delaware River, Delaware–New Jersey–Pennsylvania, and Kennebec River, Maine, populations. However, despite intensive efforts, we failed to find evidence of successful spawning of striped bass in the Kouchibouguac River in 1996 (Robinson et al. 1998) or in the Kouchibouguac or Richibucto rivers in 1997 or 1998 (Robinson et al. 2001), thus disputing the likelihood of successful striped bass spawning in these rivers.
The only spawning population identified in the southern Gulf of St. Lawrence is located in the northwest Miramichi River, the largest estuary in the southern Gulf of St. Lawrence (Robichaud-LeBlanc et al. 1996; Bradford and Chaput 1997; Bradford et al. 1999a). Striped bass spawn in the northwest Miramichi River on an annual basis (Robichaud-LeBlanc et al. 1996, 1998; Bradford and Chaput 1997; Bradford et al. 1999b). Adult mark–recapture data also support the conclusion that the Miramichi River is the major, if not the only, spawning ground for striped bass within the southern Gulf of St. Lawrence (Bradford and Chaput 1997; Bradford et al. 1999a). Adult striped bass tagged in rivers throughout the southern Gulf of St. Lawrence have been recaptured in spawning condition on the spawning grounds in the northwest Miramichi River in late spring (Bradford and Chaput 1998).
Our result suggesting that age-0 striped bass move into coastal waters of the southern Gulf of St. Lawrence is not consistent with the perception that age-0 striped bass are exclusively resident within their natal estuaries, particularly at both extremes of their Atlantic coast distribution (Rulifson and Dadswell 1995; Secor and Piccoli 1996; Wainright et al. 1996). Most of the previous research on movement of age-0 striped bass has been in the two largest U.S. spawning systems, the Chesapeake Bay and the Hudson River (Secor and Piccoli 1996; Wainright et al. 1996). Striped bass from the Chesapeake Bay under the age of 1 year are believed to remain in or close to their natal river systems (Mansueti 1961) and do not undergo migrations through moderate- to high-salinity coastal waters (Kohlenstein 1981). Similarly, age-0 striped bass from the Hudson River do not undergo extensive coastal migrations, although they potentially may range over 200 km of river from the most upriver freshwater spawning sites to lower-estuary rearing habitats. Two hundred kilometers is approximately equal to the distance between the spawning grounds in the northwest Miramichi River and the furthest upstream locales in the Richibucto River where age-0 fish were captured in this study. Additionally, a portion of the Hudson River age-0 striped bass population is known to leave the river to inhabit nearby coastal embayments within 20 km of the river mouth. Thus, age-0 striped bass from the Miramichi River may exhibit greater coastal movements through moderate-salinity waters than striped bass from other populations.
The movement of age-0 striped bass was inferred to extend as far south as the Richibucto River, and it is possible that their dispersal may extend even further south, through the Northumberland Strait towards Nova Scotia. In 1998, First Nation fishers reported large catches of age-0 striped bass from salmon trap nets on the River Philip, Nova Scotia. Catches were verified when 57 age-0 striped bass, ranging from 14.5 to 19.7 cm TL, were collected in November of 1998 (M. Robinson, unpublished data). The closest area where spawning is known to occur is the Miramichi estuary, 450 km to the north, making it a possible source of these age-0 striped bass. There are currently few data available to help determine whether age-0 striped bass also move to the north from the Miramichi estuary, although this seems possible in light of the extent of their southerly movements. The similarity in mtDNA length haplotypes between age-0 and yearling striped bass from the Miramichi River and yearling fish from the Tabusintac River to the north supports this possibility (Wirgin et al. 1993).
It is unknown whether the age-0 striped bass that utilize the Kouchibouguac and Richibucto rivers successfully overwinter in these systems. Older striped bass from the Miramichi estuary are known to overwinter in the Kouchibouguac River (Hogans and Melvin 1984; Bradford et al. 1999a) and Richibucto River (Rulifson and Dadswell 1995) above the salt front in freshwater or nearly freshwater, and in recent years, age-0 striped bass have also been observed overwintering in the Richibucto River (R. G. Bradford, Fisheries and Oceans Canada, Dartmouth, Nova Scotia, personal communication). Although age-0 striped bass were not sampled at overwintering locations in the Kouchibouguac and Richibucto rivers in 1997 and 1998, it is possible that some age-0 fish that enter these systems in late summer and grow to fork lengths exceeding 100 mm would successfully overwinter (Bernier 1996; Bradford and Chaput 1997). The movement of age-0 striped bass from the Miramichi River to overwintering sites in the Kouchibouguac and Richibucto estuaries could thus provide a mechanism for the recolonization of other southern Gulf of St. Lawrence rivers (presuming that striped bass inhabited these rivers previously).
In earlier studies, the presence or absence of age-0 striped bass has been used as an indicator of striped bass spawning in some southern Gulf of St. Lawrence rivers (Melvin 1979; Hogans and Melvin 1984). Our research has demonstrated that some rivers in the southern Gulf of St. Lawrence provide rearing habitat for age-0 striped bass, even if they are not presently supporting striped bass spawning. In light of the results presented here, we suggest that the presence of age-0 striped bass within a given river in the southern Gulf of St. Lawrence in late summer or early fall cannot be used as an indicator of spawning occurrence in that river.
Acknowledgments
We thank personnel at Kouchibouguac National Park (Eric Tremblay), the Miramichi River Environmental Assessment Committee (Harry Collins), the Richibucto River Association (Gerald Beck), G. Klassen, A. Locke, and A. St.-Hilaire. We are indebted to Grace Maxwell for her assistance throughout this study. G. Saunders kindly provided laboratory facilities. We acknowledge the support of core facilities of National Institute of Environmental Health Sciences Center Grant ES00260.