Design of Innovative Divalent Cj1621 and CjaA Multiepitope mRNA-Based Vaccine Against Foodborne Campylobacter jejuni Using In Silico Approaches
Abstract
Campylobacter jejuni is one of the main causes of gastroenteritis in human and animals worldwide. Emergence of antibiotic resistance in microorganisms increased the need to develop new types of vaccines. The present study aimed to design novel multiepitope mRNA vaccine against Campylobacter jejuni using immunoinformatics tools. For this purpose, two virulence C. jejuni proteins (Cj1621 and CjaA) were selected as antigen targets, and the appropriate epitopes were predicted using immunoinformatics tools and molecular models. Five cytotoxic T lymphocyte, six helper T lymphocyte, four linear B-cell, and one conformational B-cell epitopes were linked together with an appropriate linker, and then, adjuvant (RpfE) was attached to the construct candidate. Physiochemical, immunological, secondary, and 3D structure evaluation of the proposed vaccine showed it is immunogenic, nontoxic, nonallergic, flexible, and stable. Furthermore, docking shows that the vaccine has stable interaction with the immune receptors TLR (TLR-2 and TLR-4) and B7 (B7-1 and B7-2). Moreover, analysis of the vaccine with the MD server shows its ability to induce humoral and cellular immunity of the selected host. Overall, our findings indicate that the proposed vaccine could be a promising option against Campylobacter jejuni infection; however, further lab-based studies are needed to confirm the efficiency and safety of this vaccine.
1. Introduction
Bacterial infection is a real threat to the public health around the world in the recent years. The increasing bacterial resistance to different antibiotics has led to develop new types of vaccines [1]. C. jejuni is a Gram-negative, curved bacterium, microaerophilic, and considered the most common bacteria that cause foodborne enteritis in human and farm animals [2]. However, it is commensal in birds, especially chickens. C. jejuni causes high economic losses in human and domestic animals [3]. Interestingly, dose of infection with C. jejuni is various between human and birds. Only 500–800 bacteria can incident the infection in human [4]. Infection with C. jejuni is characterized by watery or bloody diarrhea with fever and abdominal pain [5, 6].
The efforts are continuous to develop safe and effective vaccine against C. jejuni. The normal attenuated and killed vaccine might induce autoantibodies causing Guillain–Barre Syndrome for some patients. This is due to the type of lipo-oligosaccharides on the surface of the C. jejuni cell wall [7]. Thus, subunit vaccine can be the alternative type of vaccine against C. jejuni infection [8]. Multiple epitope vaccine is a modern vaccine approach, and it allows good presentation to the antigen and no covalent binding between different branches of vaccine peptides [9]. In addition, it is safe and low cost [10]. Alkyl hydroperoxide reductase (AhpC), outer membrane protein (Omp), and Flagellar L-ring protein precursor (FlgH) are campylobacter proteins that were used as subunit vaccine against Campylobacter infection in previous studies [11–13].
The mRNA vaccines mark a significant advancement in the immunization technology by introducing a genetic material that prompts cells to produce specific proteins, triggering an immune response. Their rapid development and adaptability allow for quick responses to emerging pathogens, while the absence of live virus components enhances safety. The ability of mRNA vaccines induces humoral and cellular responses, making them effective against intracellular infections [14]. Additionally, improvement in bioinformatics and computational technologies has revolutionized drug and vaccine design. These tools enable researchers to analyze biological data, predict molecular interactions, and create novel compounds with high efficacy. This accelerates the development of treatments and vaccines, enhancing our ability to combat infectious diseases [15]. In the current study, preparation of multiepitope mRNA vaccine against C. jejuni has been performed, and construct of two important virulence genes (Cj1621 and CjaA) was generated. Cj1621 is cell membrane protein, specific in Campylobacter and expected to poses trans membrane domains and conformational epitope. It is a novel periplasmic protein and can be used as a vaccine candidate against campylobacter; it is important in C. jejuni diagnosis as well [16]. C. jejuni antigen A (CjaA) is a soluble binding protein and a part of the ABC transport system. It is extra cytoplasmic and necessary for in vivo colonization of C. jejuni [17]. Also, it is well known that amino acids are vital for campylobacter metabolism. In addition to the role of amino acids as the main source of nitrogen, they have varied attributions in physiology of this pathogen. CjaA protein could be behind carrying of cysteine and other amino acids as shown in the previous study. Further, CjaA is crucial in campylobacter colonization, especially under iron deficient conditions [18, 19].
2. Materials and Methods
The work flow of the present study is provided in Figure 1.
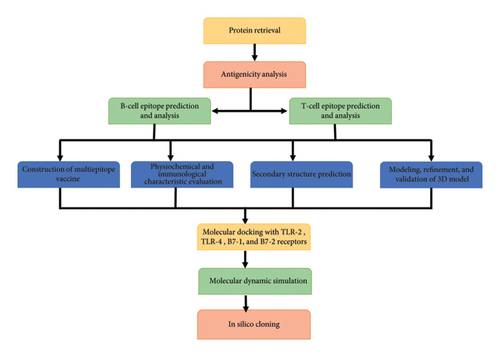
2.1. Sequence Retrieval of Protein
The UniProt Knowledgebase [20] was used for the retrieval of two selected proteins sequences of the C. jejuni NCTC 11168 strain (Cj1621 protein, accession number: Q0P807 and CjaA protein, accession number: A7H355).
2.2. Epitope Mapping
B-cell and T-cell epitopes were mapped by the IEDB server [21]. The high rank epitopes were candidate to construct of the vaccine candidate.
2.3. Human Homology
The NCBI BLASTp was used to check the homology between the predicated epitope with the homosapiens (TaxID: 9606) peptide database [22]. Epitopes with an E-value of 0.05 or higher are considered to have significant homology to the human peptide.
2.4. Evaluation of Allergenicity, Antigenicity, and Toxicity
Epitopes antigenicity was tested using VaxiJen v2.0 [23, 24]. Antigenicity was established on a 0.5 threshold. The highest antigenic epitopes were analyzed for allergenicity predictions using AllerTOP [25]. The toxicity prediction was performed by the ToxinPred web server [26].
2.5. Construction of the Multiepitope Vaccine Candidate
Epitopes were fused together to construct the proposed vaccine. Amino acid linker (GPGPG, AAY, and KK) were used to link the epitopes together. The resuscitation promoting factor E of Mycobacterium tuberculosis (UniProt ID: O53177) was selected as adjuvant and then fused to the construct via the EAAAK linker. In order to increase stability of the mRNA vaccine candidate, rabbit beta-globin and hemoglobin subunit beta (HBB) were added as UTR at C and N ends of the construct, respectively.
2.6. Physicochemical and Immunological Profile of the Vaccine Assemble
ProtParam [27] was used to estimate the physicochemical nature of the vaccine candidate. VaxiJen v2.0 and AllerTOP online tools were used to check the antigenicity and allergenicity of the vaccine construct, respectively.
2.7. Secondary Structure Prediction
The P-RoBi web server was used to predict the structural features of the proposed vaccine, such as alpha-helices, beta-turns, and regions of random coil [28].
2.8. 3D Structure Modeling Prediction, Refinement, and Validation
The 3D structure of the construct was predicted using the trRosetta server. [29]. The Galaxy Refine web server was used for refining of the proposed structure [30]. Ramachandran plot analysis was carried out using the RAMPAGE web server for the structure validation [31].
2.9. Molecular Docking
To estimate the affinity between the vaccine construct and receptor molecule, the ClusPro v2.0 server was used [32]. Also, ClusPro was applied to analyze the cooperation between the vaccine and receptor molecules. The TLR-2 (PDB ID 2Z80), TLR-4(PDB ID 2Z63), B7-1(PDB ID 1DR9), and B7-2(PDB ID 1NCN) receptors were picked and downloaded from the PDB web server.
2.10. Molecular Dynamic Simulation
Stability and motion of atoms within the complex (proposed vaccine and receptors) were predicted with the iMODS server [33], where the immune response can be elicited by the researcher via understanding dynamic behavior of the complex structures.
2.11. Vaccine 3D Assembly Flexibility
The CABS Flex 2.0 [34] which employs coarse-grained simulations to investigate protein dynamics was utilized to evaluate the structurally stable conformation of the proposed vaccine. Default parameters were applied for the distance restraint generator throughout the assessment.
2.12. Codon Optimization and mRNA Structure
Codon optimization is essential for efficient gene expression inside the foreign host. The JCat was used for codon optimization of the proposed vaccine [35]. Depending on guanine–cytosine (GC) content and the codon adaptation index, optimization of codon was generated. The secondary structure was predicted using the RNAfold server, providing insights into stability and function. In silico cloning was performed using the Snap Gene 4.2 tool [36].
3. Results
3.1. Sequence Retrieval and Antigenicity of the Vaccine Model
The selected proteins are important for colonization and amino acid metabolism in C. jejuni. Antigenicity of these proteins was estimated using the VaxiJen v2.0 server. Determination of the antigenicity was depended on a 0.5 threshold. The results show that the proposed vaccine is probable antigen with a 0.73 antigenic score.
3.2. Epitope Mapping
Only the antigenic, no allergic, nontoxic, and 100% conserved epitopes of Cj1621 and CjaA were selected, as shown in Tables 1, 2, and 3. In Table 1, five immunogenic, nontoxic, and nonallergenic cytotoxic T lymphocyte (CTL) epitopes were selected from Cj1621 and CjaA sequences to build up the candidate vaccine. Also, Table 2 shows that six helper T lymphocyte (HTL), four linear linear B-lymphocyte (LBL) epitopes (Table 3), and one B-conformational epitope were used to build the proposed vaccine (Table 4).
| Sr. no. | Epitopes | Antigenicity score | Toxicity | Allergenicity | Name of the gene | Conservancy (%) |
|---|---|---|---|---|---|---|
| 1 | FVAGVVFNV | 0.6837 | No | None | Cj1621 | 100 |
| 2 | LQVNANTNI | 1.0954 | No | None | Cj1621 | 100 |
| 3 | GVVFNVEFL | 1.1249 | No | None | Cj1621 | 100 |
| 4 | QTPQRAEQV | 0.7062 | No | None | CjaA | 100 |
| 5 | YDETLKAHF | 0.9790 | No | None | CjaA | 100 |
| Sr. no. | Epitopes | Antigenicity score | Toxicity | Allergenicity | Name of the protein | Conservancy (%) |
|---|---|---|---|---|---|---|
| 1 | EPNFSYKDGIQTPKG | 0.7560 | No | None | Cj1621 | 100 |
| 2 | LEPNFSYKDGIQTPK | 1.1344 | No | None | Cj1621 | 100 |
| 3 | LKNLQVNANTNIAPR | 0.9771 | No | None | Cj1621 | 100 |
| 4 | NQGYDIALAKRIAKE | 0.4231 | No | None | CjaA | 100 |
| 5 | NNQGYDIALAKRIAK | 0.4689 | No | None | CjaA | 100 |
| 6 | SPYMKVALGVAVPKD | 0.4887 | No | None | CjaA | 100 |
| No. | Start | End | Peptide | Length | Gene name | Toxicity |
|---|---|---|---|---|---|---|
| 1 | 28 | 45 | FLGKKDSNHDLYFSRNIE | 18 | Cj1621 | Nontoxin |
| 2 | 62 | 80 | WANENLSSKITLDNSEKAE | 19 | Cj1621 | Nontoxin |
| 3 | 23 | 36 | NSDSKTLNSLDKIK | 14 | CjaA | Nontoxin |
| 4 | 49 | 54 | KPPFGY | 6 | CjaA | Nontoxin |
| Residue | Location | Score |
|---|---|---|
| YKDGIQTPKGAAYLEPNFSYKDGIQTPKAAYLKN | 76–109 | 0.752 |
3.3. Construction of the Proposed Vaccine
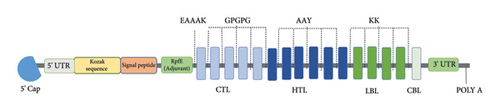
The vaccine candidate generates utilizing CTL, HTL, and LBL epitopes that were mentioned previously. As shown in Figure 2, the final construction of the mRNA vaccine candidate includes 5 m7GCap, HBB, Kozak sequence, SP, amino acid linker, RpfE, 15 B and T-cell epitopes, rabbit beta-globin, and poly A tail (Figure 2). AAY, GPGPG, and KK were used to link up the epitope’s sequences together.
3.4. Physiochemical and Immunological Analysis of the Vaccine
VaxiJen, ANTIGENpro, and AllerTOP server were applied to estimate the immunogenicity of the candidate. Results show that the vaccine candidate is antigenic, nonallergic, and nontoxic to the host cell. The designed protein has a molecular weight of 26,198.64 Da; the chemical formula is C1719H2657N477O515S2; estimated half live in the mammalian cell is about 1.9 h, in bacteria is 10 h, and in yeast is 20 min in vivo. The theoretical isoelectric point is 9.51, and the grand average of hydropathicity is −0.587 (Table 5). Overall, the results reveal that the proposed vaccine is immunogenic and stable.
| Properties | Measurement | Note |
|---|---|---|
| AA | 357 | Suitable |
| MW | 26,198.64 Da | Appropriate |
| Theoretical pI | 9.51 | Basic |
| Formula | C1719H2657N477O515S2 | — |
| Half-life (Escherichia coli) | 10 h | — |
| Half-life (mammalian reticulocytes) | 1.9 h | — |
| Half-life (yeast cells) | 20 h | — |
| Grand average of hydropathicity | −0.587 | Hydrophilic |
| Instability index of the proposed vaccine | 25.91 | Stable |
| Antigenicity | 0.7381 | — |
| Allergenicity | Nonallergic | — |
| Toxicity | Nontoxic | — |
3.5. Secondary Structure of the Proposed Vaccine
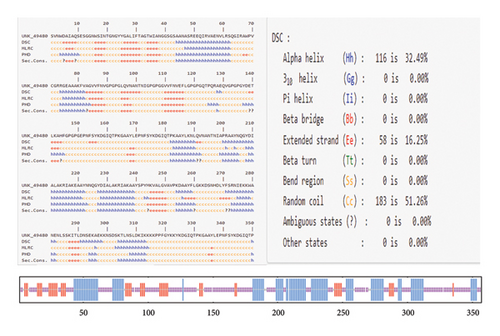
The secondary structure of the vaccine was predicted using the PRABI server. The constructed protein, comprising 357 amino acids, was found to contain 32.49% alpha helices, 16.25% extended strands, and 51.26% random coils (Figure 3). It was noticed that during the infection, the unfolded regions in the protein are detected by the antibodies.
3.6. Modeling, Refinement, and Validation of the 3D Model
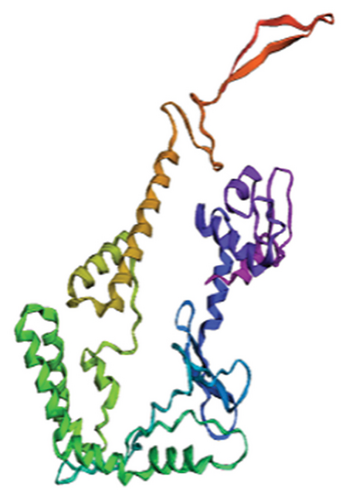
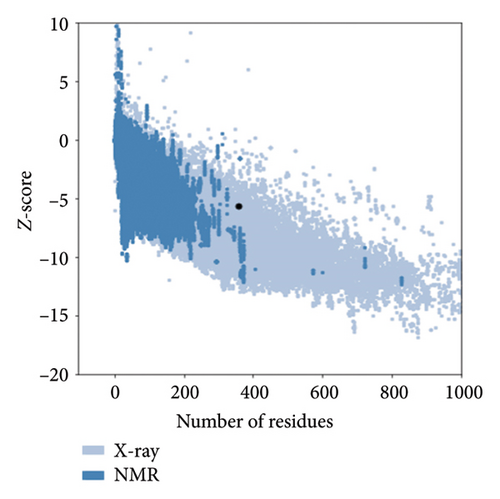
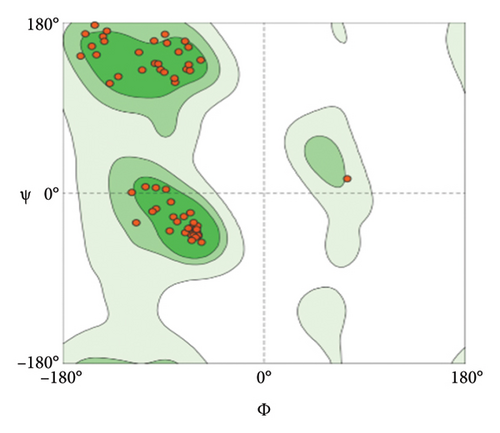
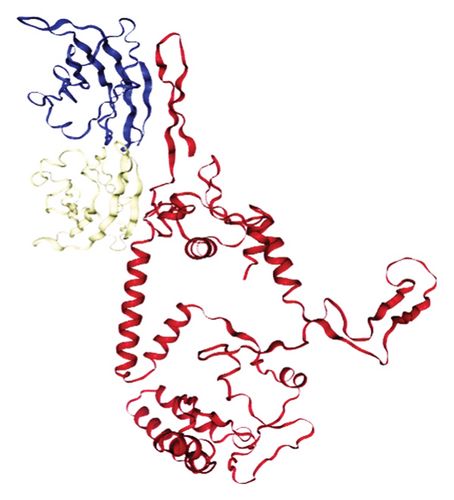
Out of the 5 models, Model 1 was selected as the tertiary structure of the proposed vaccine using trRosetta (Figure 4(b)). The Galaxy web tool was used to refine the selected structure (Figure 4(c)). The Z score that was estimated with ProSA was −5.68, while Ramachandran favored and outliers were 97.26% and 0.35, respectively, in SWISS-MODEL software simulation (Figures 4(d), 4(e), and 4(f)).
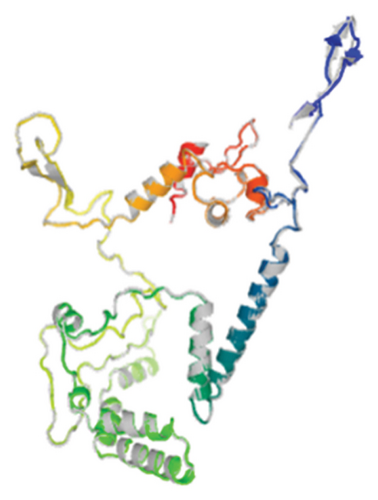
3.7. Molecular Docking and Molecular Dynamic Simulation
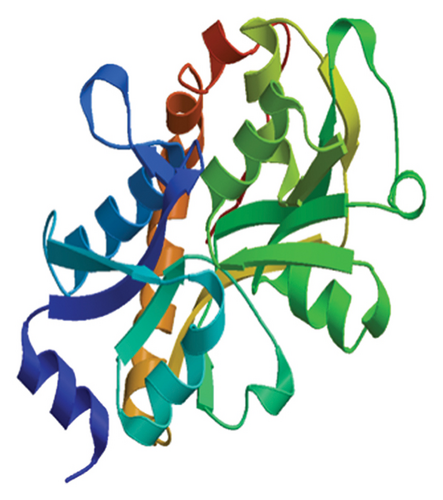
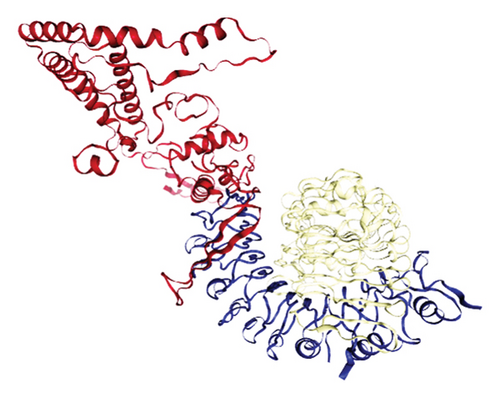
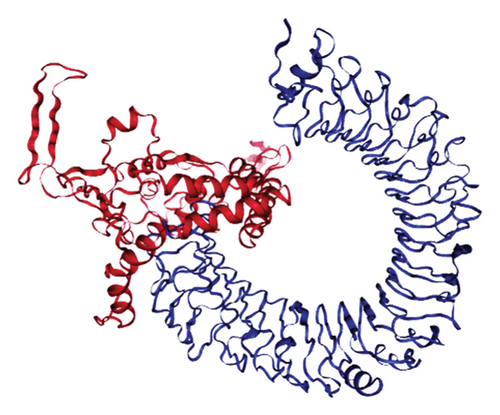
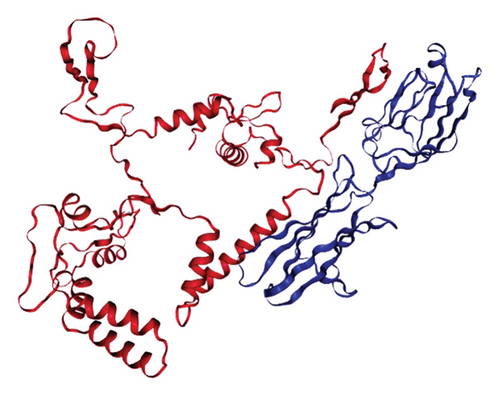
The results of the server (LZerD web server) showed that there are 10 possible models between the proposed vaccine and the TLR2, TLR4, B7-1, and B7-2 receptors. Based on the sum-rank score, the most stable model has been chosen (Table 6, Figure 5).
| Receptor | GOAP score | GOAP rank | DFIRE score | DFIRE rank | ITscore score | ITscore rank | Score RankSum |
|---|---|---|---|---|---|---|---|
| TLR2 | −118570.90 | 40 | −86935.85 | 90 | −42791.22 | 11 | 141 |
| TLR4 | −114654.84 | 40 | −80830.43 | 17 | −40608.73 | 6 | 63 |
| B7-1 | −54628.14 | 124 | −38140.12 | 34 | −20752.52 | 5 | 163 |
| B7-2 | −55861.26 | 168 | −40647.57 | 94 | −22225.32 | 27 | 289 |
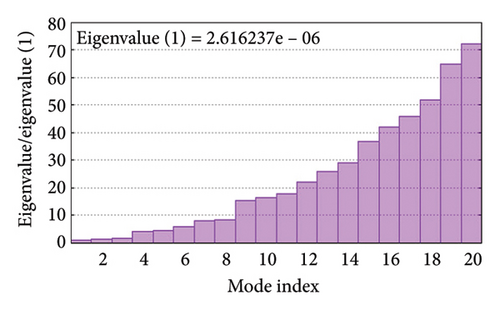
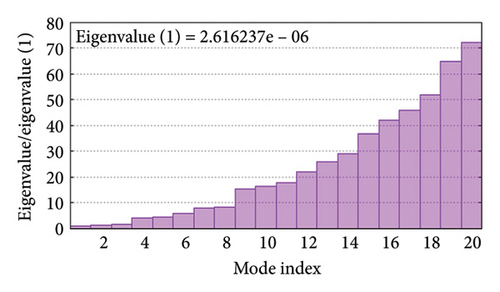
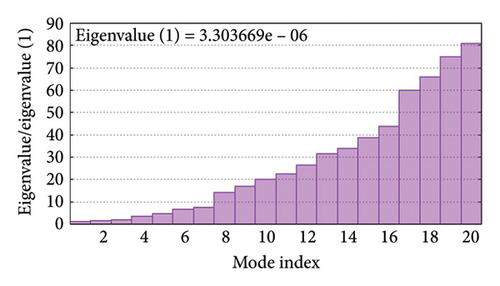
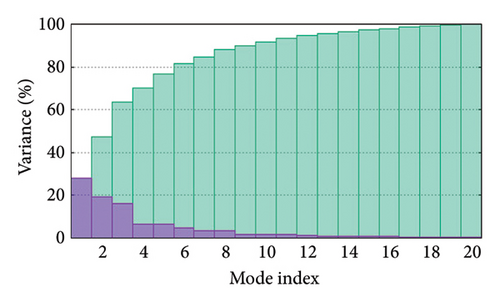
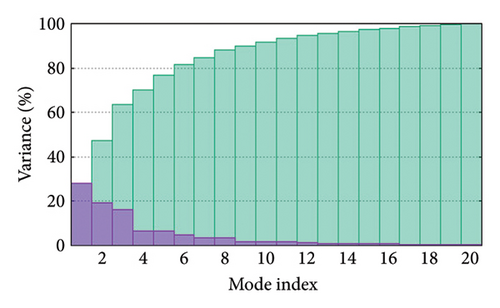
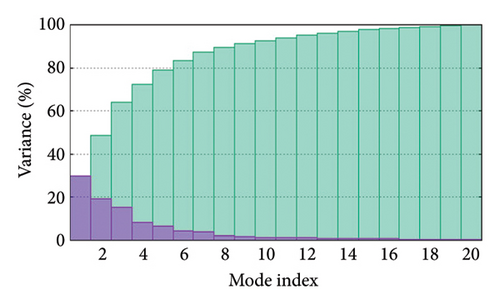
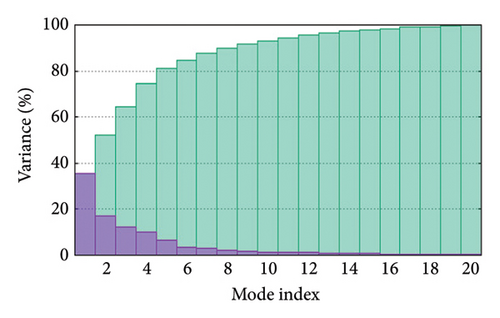
As shown in Figure 5, eigenvalues of the docked complexes were 2.61623437e−6, 2.2616237e−6, 5.303669e−6, and 2.233684e−6. These results indicate that four complexes have strong deformability. In Figure 6(b), the pure bar refers to the individual variation, while green bar represents accumulative variation. Analysis of the variance plot shows that the individual variance of successive mode exhibited a modest decline.

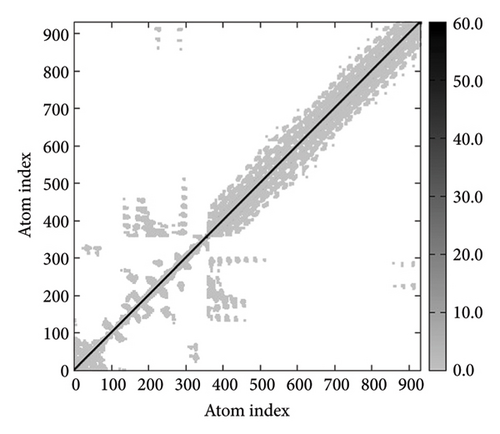
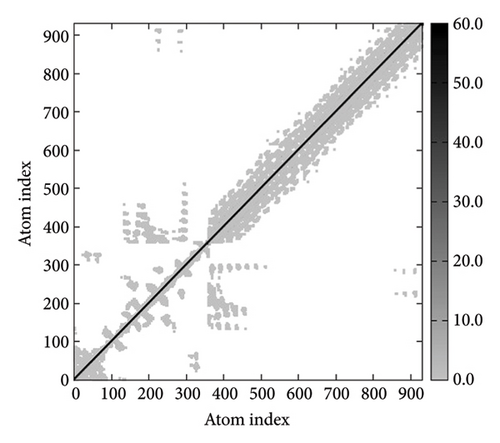
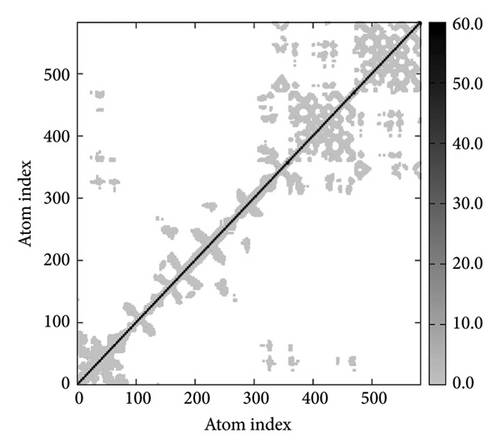
In Figures 7(a), 7(b), 7(c), and 7(d), the degree of correlation within amino acid duplets scattered in dynamical regions was represented in different colors. The red color refers to correlated residue, and the blue color refers to noncorrelated residues, while white color representing anticorrelated amino acid duplets. In addition, the elastic network model that formed by combination of the vaccine with the receptors was generated to differentiate the atom pairs connected by springs (Figure 7(e), 7(g), 7(h)). Thus, the above results indicate that the vaccine-receptor complex has good stability and stiffness.
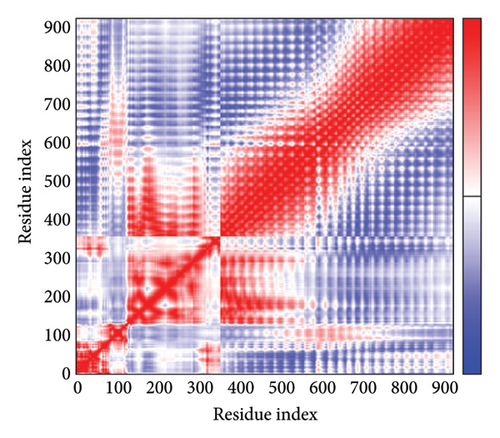
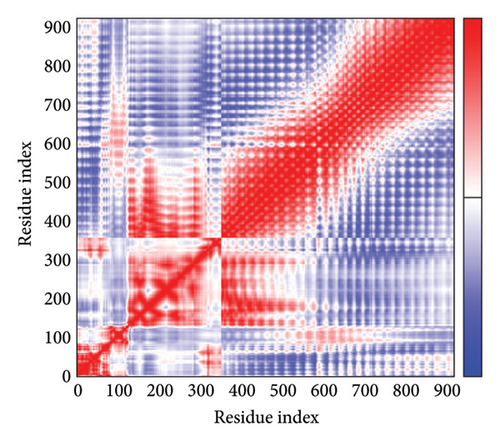
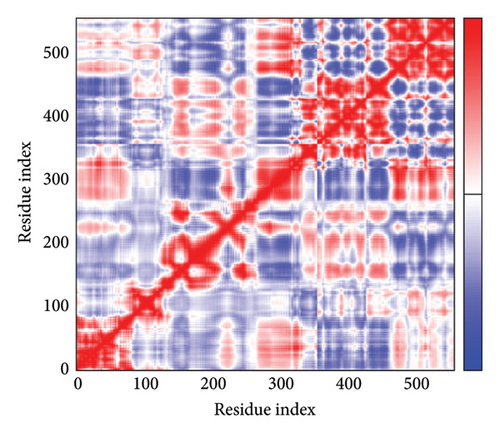
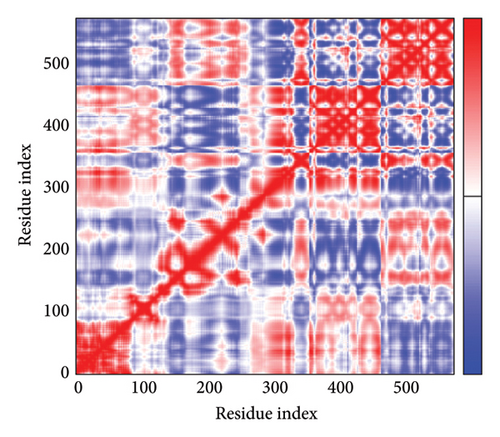

3.8. Immunostimulation
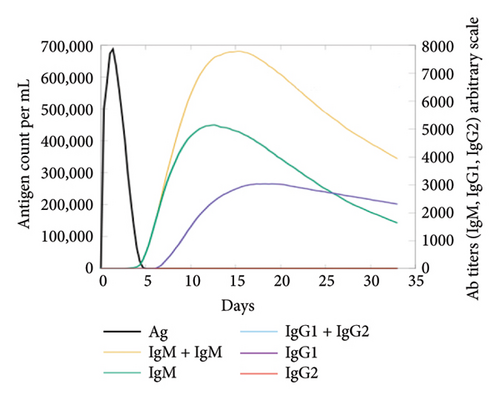
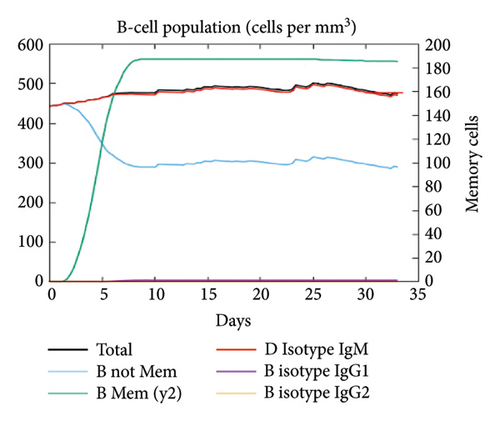
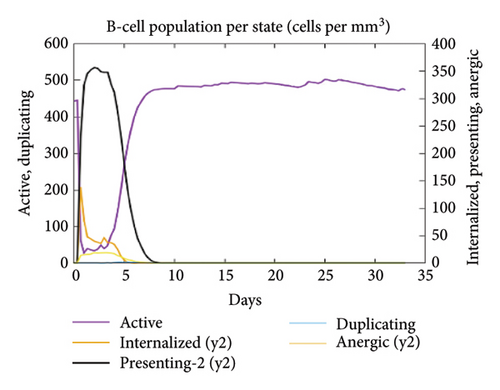
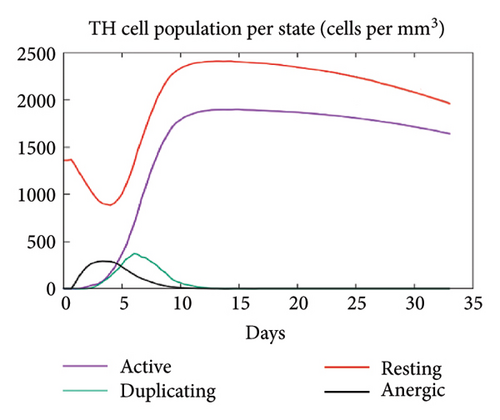
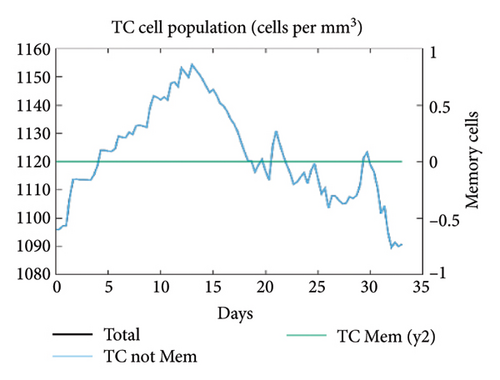
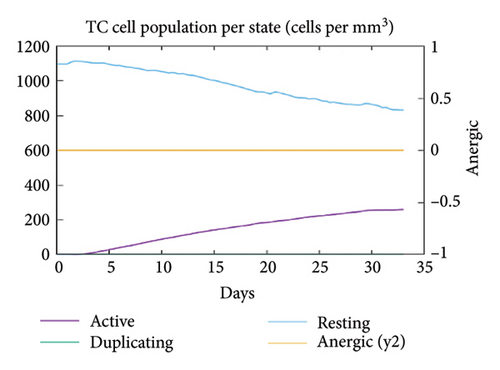
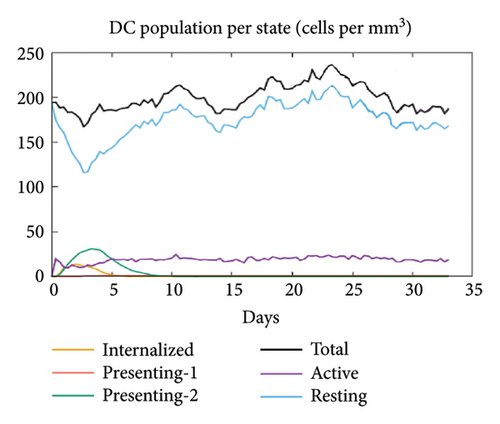
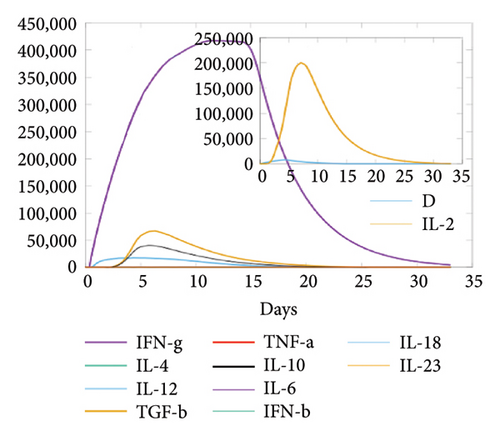
In the current study, predication of the immune response to three injections of the vaccine was performed using the C-ImmSim server (Figure 8(a)). In the 2end and 3ride injections of the vaccine, the concentrations of immunoglobulin M (IgM) and IgG continued to rise. B cells were observed and involved in the immune response, specifically in the humoral type. As shown in Figures 8(b) and 8(c), the population of B cells increases with three doses of the modeled vaccine. In the same manner, T cells (helper and cytotoxic) were also increased (Figures 8(d), 8(e), and 8(f)). In Figure 8(g), macrophages were enhanced after three doses of the vaccine, while dendritic cells remained stable (Figure 8(h)). Levels of IFN-γ and IL-2 cytokines were increased (Figure 8(i)). Finally, the low level of the Simpson index (D) refers to the diverse immune response. Overall, these findings indicated that the mRNA multiepitope vaccine induces a good immune response against C. jejuni.
3.9. Structural Versatility Assessment
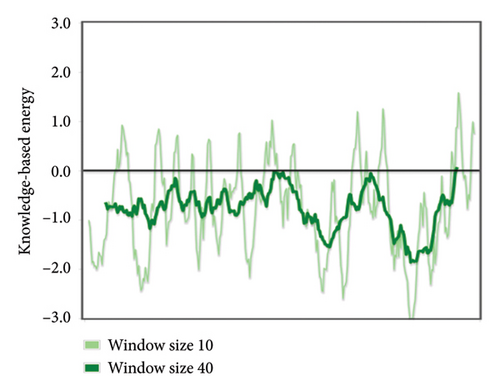
The structural versatility of the formulated vaccine was evaluated using CABS-flex, a modeling technique known for its rapid simulation abilities. The results presented in this section demonstrate that a substantial number of protein residues display favorable stability. The fluctuation profile of residues in the engineered protein vaccine has been illustrated in Figure 9 for reference.
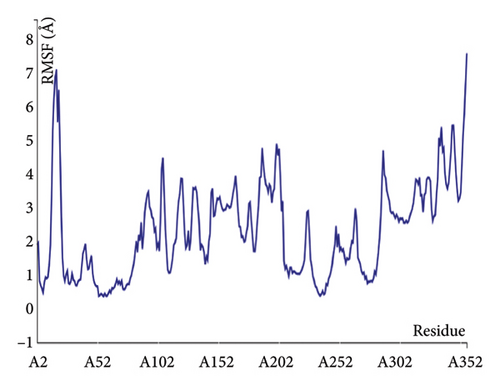
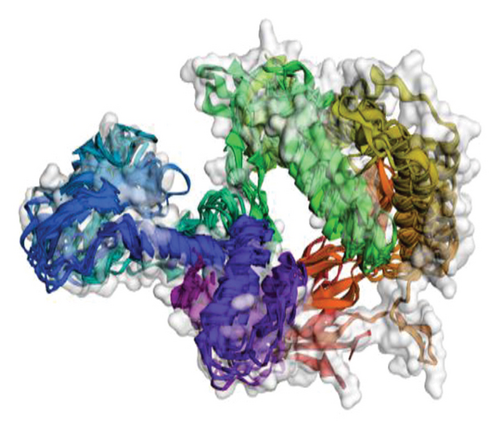
3.10. mRNA Structure and Computerized Cloning
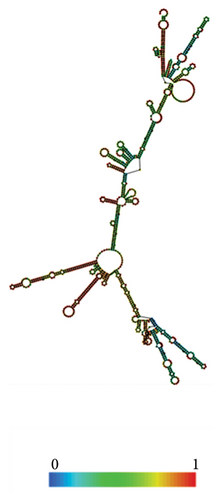
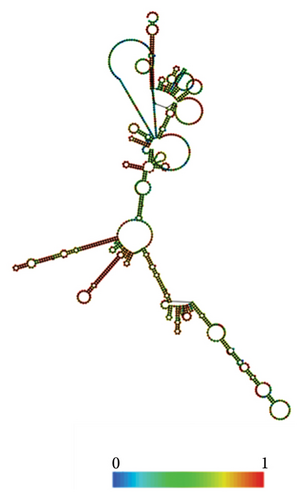
The RNAfold server was used to predict the structural arrangement of the mRNA (Figures 10(a), 10(b)). The results demonstrate that the mRNA vaccine is expected to exhibit stability, as evidenced by MFE of −419.90 kcal/mol for the structure. Additionally, the secondary centroid structure displayed a free energy of −329.50 kcal/mol (Figure 10(a)). The free energy of the thermodynamic ensemble was −440.61 kcal/mol (Figure 10(b)). These findings provide valuable insights into the thermodynamic robustness of the mRNA vaccine structure.
Result of codon optimization shows that GC content of the proposed fragment was 68.2%, while CAI was 0.95. As shown in Figure 11, the proposed fragment was cloned in the vector (pET21) at position 198 and 1343 at the position of the selected restricted sites (EcoRI and BamHI).
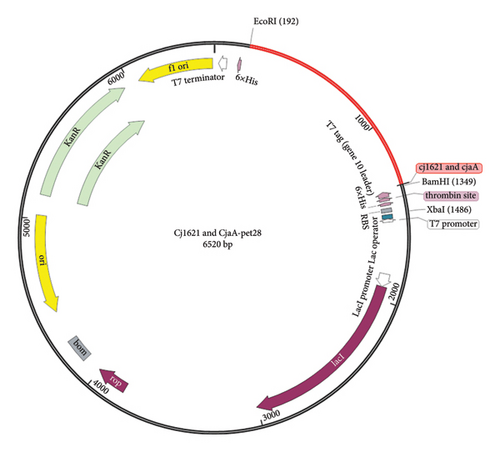
4. Discussion
Using of in silico approaches to find multiple epitope vaccine is the effective way to control the increasing antibiotic resistance in pathogenic bacteria. The findings that presented in this study highlight significant advancements in the design of the multiepitope mRNA vaccine candidate against C. jejuni. The selected proteins, Cj1621 and CjaA, were evaluated for their antigenicity, epitope mapping, and subsequent construction of multiepitope vaccine. This demonstrates promising characteristics that warrant further exploration [37–40]. The VaxiJen v2.0 server’s antigenicity assessment revealed strong recognition potential by the immune system, highlighting the viability of these candidates as potential vaccines. A thorough epitope analysis identified 15 immunogenic epitopes that are nontoxic and nonallergenic. These epitopes are capable to induce both B and T cell response, including 5 CTL epitopes, 6 HTL epitopes, 4 LBL epitopes, and one conformational B epitope. This diverse selection enhances the potential for a robust immune response.
The multiepitope vaccine was constructed with critical elements such as m7GCap, UTR (HBB), Kozak sequence, signal peptide, adjuvant (RpfE), and flexible linkers (EAAAK). This design promotes optimal folding and presentation of the epitopes. The physicochemical evaluation indicated a stable protein with a molecular weight of approximately 26,198.64 Da and a PI of 9.51, which suggests solubility under physiological conditions. Furthermore, estimated half-lives in various environments indicate vaccine’s sustained effectiveness in mammalian cells, bacteria, and yeast. These findings collectively highlight the potential of our multiepitope vaccine candidate for advancing protective strategies against C. jejuni infections. Immunological analysis further confirmed that the candidate is nontoxic, nonallergic, and antigenic. These characteristics are essential for ensuring safety and efficacy in potential clinical applications. The stability and favorable properties of the chimeric protein underscore its suitability as a vaccine candidate. The 3D model of the vaccine construct was developed using trRosetta and refined with the Galaxy web server, resulting in a stable model with positive validation outcomes. Validation through SWISS-MODEL and ProSA confirmed the reliability and accuracy of the model for future studies. Moreover, molecular docking simulations with B7-1, TLR2, TLR4, and B7-2 receptors demonstrated the strong binding and stability of the complexes. The molecular dynamics analysis further supported the stability of these complexes, highlighting the deformability and favorable structural integrity of the interactions, essential for inducing an effective immune response. The immunostimulation analysis using the C-ImmSim server revealed a promising immune response elicited by the modeled vaccine and the increased amount of IgG and IgM antibodies. Also, proliferation of B and T cells indicates the ability of the vaccine to trigger both type of immune response (humoral and cellular). Additionally, the upregulation of cytokines and macrophages underscores the immunomodulatory properties of the vaccine construct. The CABS-flex analysis revealed that a substantial portion of protein residues in the engineered vaccine exhibits favorable stability, indicating promising structural characteristics crucial for vaccine efficacy. Moreover, the mRNA vaccine’s structural prediction utilizing the RNA fold server indicated significant stability, evident by the minimum free energy values obtained. These complementary analyses offer a comprehensive understanding of the structural versatility and stability of both vaccine formulations, enhancing our insight into their potential effectiveness. Furthermore, the computerized cloning analysis demonstrated the suitability of the vaccine construct for good expression within the selected host, as evidenced by the favorable GC content and CAI-value. The presence of EcoRI and BamHI sites for in silico cloning further facilitates the experimental validation and potential translation of the vaccine design into a practical setting. In this scientific domain, numerous studies have been carried out on a diverse range of infectious agents. The advancement of immunoinformatics and the advent of mRNA-based vaccines have led to the development of vaccines targeting pathogens such as Yersinia, Brucella, Clostridium, Mycobacterium, Pseudomonas, Toxoplasma, Listeria, and others [41–47]. The field of mRNA-based vaccine research has seen significant growth in response to these technological innovations, fostering new possibilities for combating infectious diseases.
5. Conclusion
In conclusion, the integrated computational approach utilized in this study not only provides valuable insights into the design and evaluation of a novel multiepitope mRNA vaccine but also lays the groundwork for potential experimental validation and clinical applications. This could be promising for a new avenue to combat C. jejuni infection [48].
Conflicts of Interest
The authors declare no conflicts of interest.
Funding
No funding was received for this manuscript.
Acknowledgments
The authors would like to thank the Deanship of Veterinary Medicine College at Al-Qadisiyah University for supporting this work.
Open Research
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.




