Genetic Predictive Association Between Inflammatory Cytokines and Vitiligo: A Bidirectional Mendelian Randomization Study
Abstract
Background: Previous studies have indicated a close association between vitiligo and inflammatory cytokines, yet the genetic predictive association remains unclear. The study aimed to predict the relationship between vitiligo and inflammatory cytokines in genetics.
Methods: A bidirectional analysis was conducted by the two-sample MR. We selected independent genetic tools for significant levels associated with inflammatory cytokines and vitiligo from corresponding meta-analyses of genome-wide association study (GWAS). The summary data for inflammatory cytokines were derived from a GWAS meta-analysis conducted on three separate Finnish population cohorts, comprising a total of 8293 individuals (N = 8293). The vitiligo data, on the other hand, were extracted from a GWAS meta-analysis conducted on individuals of European ancestry, totaling 44,266 individuals (N = 44,266). As the primary analytical approach, we employed the inverse variance weighted (IVW) method, and to ensure the reliability of our main findings, we also performed sensitivity analysis to evaluate the robustness.
Results: Compelling evidence indicated a causal association between genetically predicted IL-18 and IL-8 with vitiligo (odds ratio, OR: 1.162, 95% CI: 1.043–1.295, p = 0.006, and pfdr = 0.020 and OR: 0.726, 95% CI: 0.602–0.876, p = 0.001, and pfdr = 0.004). In addition, cytokines including stromal cell-derived factor-1 alpha (SDF1a) (Beta: 0.032, p = 0.016, and pfdr = 0.039), IL-17 (Beta: 0.038, p = 0.005, and pfdr = 0.027), basic fibroblast growth factor (FGFBasic) (Beta: 0.028, p = 0.034, and pfdr = 0.170), monocyte chemoattractant protein-1(MCP1) (Beta: 0.026, p = 0.046, and pfdr = 0.189), and platelet-derived growth factor BB (PDGFbb) (Beta: 0.026, p = 0.040, and pfdr = 0.198) were suggested to be the consequences of vitiligo.
Conclusions: According to this investigation, it has been proposed that the etiology of vitiligo is likely associated with IL-18 and IL-8, whereas the downstream progression of vitiligo can be attributed to several inflammatory cytokines such as IL-17, SDF1a, PDGFbb, MCP1, and FGFBasic.
1. Introduction
Vitiligo is a prevalent systemic skin disorder characterized by the idiopathic destruction of melanocytes, leading to a loss of pigmentation. The main manifestations of the skin lesions are depigmented spots of varying sizes. Globally, its prevalence ranges from 0.5% to 2% [1]. While the precise etiology remains elusive, current understanding posits connections to autoimmune responses, cytokine activity, melanocyte autophagy, neuropsychiatric, genetic, oxidative stress, and environmental factors [2]. Vitiligo is now recognized primarily as an autoimmune disorder, with the disruption of the immune system and abnormalities in inflammatory factors playing crucial roles in its pathogenesis. Although frequently perceived as a mere cosmetic concern, vitiligo significantly impairs the patient’s quality of life and mental wellbeing, thereby perpetuating the disease’s deleterious cycle [3, 4].
In the immunological cascade implicated in vitiligo, numerous cytokines contribute to its pathogenesis. A comprehensive review pinpointed biomarkers correlating with vitiligo activity in both skin and blood and assessed their utility based on sensitivity and specificity [5]. Present evidence strongly supports IL-1β, IL-17, IFN-γ, TGF-β, sCD25, autoantibodies, and oxidative stress markers as pertinent circulating biomarkers. Chemokines, owing to their robust expression, also present as biomarkers. In the foreseeable future, CXCL9, CXCL10, and CXCL12 might gain traction in clinical evaluations. IL-17 fosters leukocytes and melanocyte interactions. The proinflammatory Th17 cells, which infiltrate the dermis in advanced vitiligo, produce IL-17 and IL-17F. IL-17, in concert with local inflammatory mediators such as IL-1β, IL-6, and TNF-α, curtails melanocyte proliferation [6]. Exposing melanocytes to TNF-α externally markedly diminishes their viability and augments cellular and mitochondrial reactive oxygen species levels [7]. Notably, the early stages of vitiligo see the migration of T-cells into the skin facilitated by MIG/IP10/CXCR3 axis driven by interferon (IFN) IFN-γ. Elevated circulating MIG and IP10 levels characterize progressive vitiligo [8].
The role of the inflammatory response in vitiligo remains a subject of debate: is it a cause of vitiligo or does it arise from disease progression with subsequent alterations in inflammatory factor levels? While numerous studies have probed the link between inflammatory cytokines and vitiligo, their conclusions might be obfuscated by confounding factors or potential reverse causality, making the results elusive [9–12].
Mendelian randomization (MR) is a recognized tool in genetic epidemiology for etiological inference [13]. As the MR research method has matured, it is been adopted to discern the pathogenesis between complex diseases. In this study, using genome-wide association study (GWAS) data of 41 kinds of inflammatory cytokines and vitiligo, we aim to unveil the genetic association between these cytokines and vitiligo using the two-sample MR method, thereby informing potential vitiligo treatment strategies in clinical practice.
2. Materials and Methods
This study involved human participants, and each participant signed a written informed consent form, which received ethical review and approval as it met the requirements of local legislation and institutions. Due to the use of publicly available, anonymized, and deidentified GWAS aggregated data, this study was exempt from additional ethical review approval.
2.1. Study Design
In this study, a two-sample MR analysis was conducted to explore the potential genetic predictive relationship between inflammatory cytokines and vitiligo. The associated data for exposure and outcomes were obtained from the GWAS database. MR research should meet the following basic assumptions: (i) instrumental variables (IVs) are directly related to exposure; (ii) IVs have no direct correlation with outcome but only affect outcome through exposure; and (iii) IVs are not correlated with confounding factors. The study design process was illustrated in Figure 1.
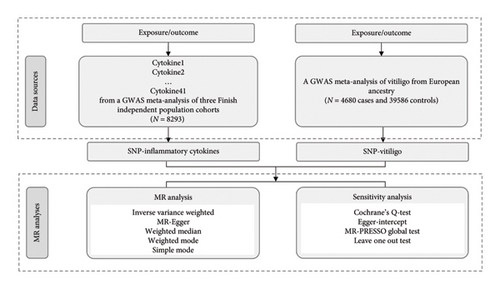
2.2. Data Sources
The assembled information in this investigation was derived from openly accessible GWAS databases. The GWAS meta-analysis of three independent Finnish population cohorts, comprising 8293 individuals from Finland, produced data regarding 41 cytokines and growth factors [14]. Regarding vitiligo, the GWAS data were extracted from a GWAS meta-analysis enlisted in the GWAS Catalog (GCST004785) [15]. The analysis involved 4680 vitiligo cases and 39,586 controls with European ancestry. The aforementioned GWAS data originated from European populations. To our understanding, there exists no overlap of samples between exposure and outcome GWASs.
2.3. Selection of Genetic IVs
When vitiligo was used as an exposure factor, we selected single-nucleotide polymorphisms (SNPs) from the data that were significantly related to vitiligo (p < 5 × 108). When inflammatory cytokines were used as exposures, a slightly relaxed threshold (p < 5 × 10−6) was applied to include more SNPs. In order to eliminate the bias caused by the linkage disequilibrium, the SNPs must satisfy the parameters r2 < 0.001 and a clumping window greater than 10,000 kb. In addition, the F-statistic, indicating the strength of the correlation between SNPs and exposure factors, was required to be greater than the conventional value of 10 [16].
2.4. Statistical Analyses
The methods employed included inverse variance weighted (IVW) [17], MR-Egger [18], weighted median, weighted mode, and simple mode. The IVW method was the main analysis method, supplemented by other analysis methods. Furthermore, the Benjamini–Hochberg method was employed to rectify the p values obtained from conducting multiple tests. A false discovery rate (FDR) lower than 0.05 signifies statistical significance and provides compelling result. If the p value was less than 0.05 before correction, but the FDR was greater than 0.05, the results were considered no longer significant after FDR correction. To further explain the potential pleiotropy, a series of sensitivity analyses were performed. Cochrane’s Q test quantified the heterogeneity of IVs. As a weighted linear regression, MR-Egger intercept can evaluate whether there is horizontal pleiotropy of IVs. The MR-PRESSO technique is capable of identifying and rectifying abnormal SNPs by examining the remaining sum and assessing for outliers. Furthermore, the leave-one-out sensitivity analysis was utilized to determine whether a single SNP had a substantial impact on the causal effect. The aforementioned procedures were conducted utilizing R Software 4.3.1 (https://www.R-project.org) through the implementation of TwoSampleMR (0.5.7 Version), MendelianRandomization (0.8.0 Version), and MRPRESSO package (1.0.0 Version).
3. Results
3.1. Influence of 41 Inflammatory Cytokines on Vitiligo
Genetically determined levels of 41 inflammatory cytokines were analyzed to investigate their potential association with vitiligo. The study utilized a range of 3–17 SNPs, with explained variances ranging from 1.8% to 20.7%. To avoid bias from weak IVs, F-statistic values were carefully considered and ranged from 11.16 to 789.15 (Table S1). It is demonstrated that the IVs included in the MR analysis have sufficient strength to confirm the reliability of the results.
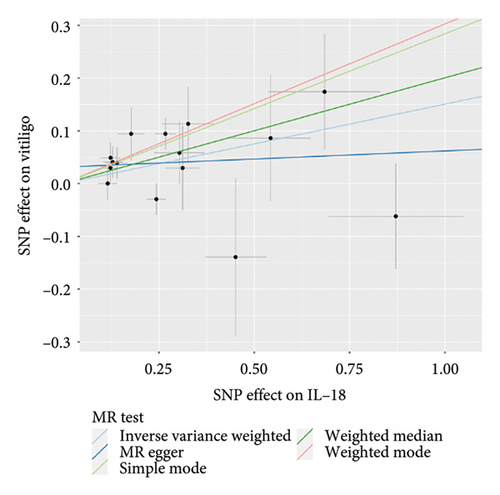
The main results of MR analysis of influence of 41 genetically predictive inflammatory cytokines on vitiligo were shown in Figure 2. In this MR analysis, IVW, the primary analysis method, showed two significant genetic predictive relationships with FDR-corrected p values less than 0.05. Specifically, genetically predicted IL-18 and IL-8 have been linked to vitiligo risk, showing different effects. Figure 3 presented the scatter plots and funnel plots depicting the MR analyses conducted on IL-18 and IL-8 in vitiligo. The IVW analysis demonstrated a significant association between genetically predicted IL-18 and the development of vitiligo (odds ratio, OR: 1.162, 95% CI: 1.043–1.295, p = 0.006, and pfdr = 0.020). The gene predicted that for every additional standard deviation unit of IL-18, the risk of developing vitiligo increased by 16.2%. The weighted median analysis method was consistent with the IVW analysis method (OR: 1.222, 95% CI: 1.053–1.417, p = 0.008, and pfdr = 0.020), further proving the genetic predictive relationship between IL-18 and vitiligo. However, MR-Egger (OR: 1.031, 95% CI: 0.841–1.264, p = 0.772, and pfdr = 0.772), weighted mode (OR: 1.354, 95% CI: 0.920–1.991, p = 0.146, and pfdr = 0.183), and simple mode (OR: 1.328, 95% CI: 1.011–1.745, p = 0.061, and pfdr = 0.101) results were not significant.
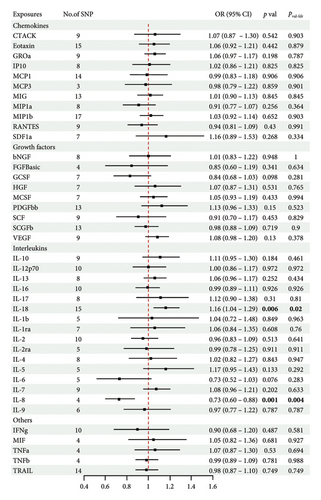
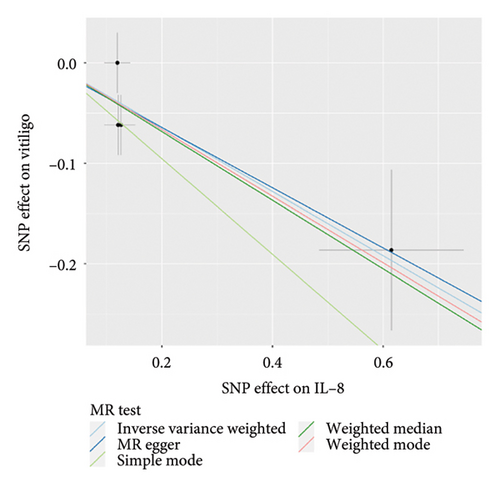
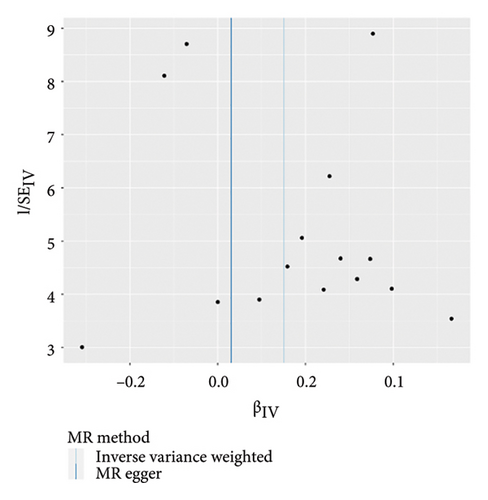
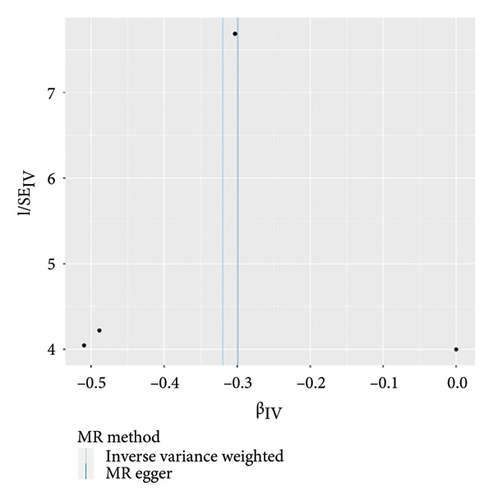
On the contrary, a 27.4% reduction in the risk of vitiligo was observed for every standard deviation unit increase in genetically predicted IL-8 (OR: 0.726, 95% CI: 0.602–0.876, p = 0.001, and pfdr = 0.004). The weighted median (OR: 0.711, 95% CI: 0.551–0.917, p = 0.009, and pfdr = 0.021) supported this finding. However, MR-Egger, weighted mode and simple mode did not yield significant results (p > 0.05).
In addition, there was no genetic predictive relationship between the other 39 inflammatory cytokines and vitiligo at the genetic level. Moreover, the results of the other four analysis methods showed similar trends (Table S2). As only three SNPs were significantly and independently associated with monocyte-specific chemokine 3 (MCP3), the IVW method was exclusively applied.
In the sensitivity test, the analysis did not reveal any evidence supporting heterogeneity in the associations between IL-18 and IL-8, as assessed by Cochrane’s Q statistic (p = 0.154 and p = 0.432). As part of further analysis, we conducted the MR-PRESSO global test and identified a single outlier with regards to cutaneous T cell-attracting chemokine (CTACK) (rs10474392).
After removing the outlier, we re-examined the analysis. No pleiotropy was detected in the association of 41 genetically predictive inflammatory cytokines and vitiligo (p > 0.05). Meanwhile, the MR-Egger intercept did not detect any potential horizontal pleiotropy (p > 0.05), indicating that there were no confounding factors affecting our results (Table S3). Leave-one-out analysis showed that MR analysis results did not fluctuate due to the exclusion of a single SNP. Figure S1 displayed forest plots and leave-one-out sensitivity analyses of MR analyses conducted for IL-18 and IL-8 in vitiligo.
3.2. Influence of Vitiligo on 41 Inflammatory Cytokines
With vitiligo as an exposure factor and inflammatory cytokines as outcomes, we extracted 34 SNPs as IVs for vitiligo. The explained variances were 82.63% and F-statistic values were considered and ranged from 287.59 to 3232.61 (Table S4).
The genetic predictive association between vitiligo and inflammatory cytokines was depicted in Figure 4 through MR analysis. The IVW method findings suggested a significant correlation between vitiligo and an increased level of stromal cell-derived factor-1 alpha (SDF1a) (Beta: 0.032, 95% CI: 0.006–0.058, p = 0.016, and pfdr = 0.039), IL-17 (Beta: 0.038, 95% CI: 0.011–0.065, p = 0.005, and pfdr = 0.027), basic fibroblast growth factor (FGFBasic) (Beta: 0.028, 95% CI: 0.002–0.055, p = 0.034, and pfdr = 0.170), monocyte chemoattractant protein-1(MCP1) (Beta: 0.026, 95% CI: 0.000–0.051, p = 0.046, and pfdr = 0.189), and platelet-derived growth factor BB (PDGFbb) (Beta: 0.026, 95% CI: 0.001–0.052, p = 0.040, and pfdr = 0.198). Scatter plots of these five associations between vitiligo and inflammatory factors were shown in Figure 5.
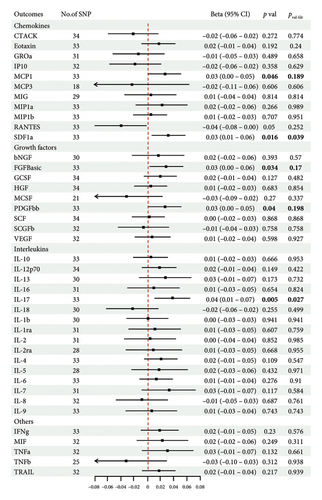
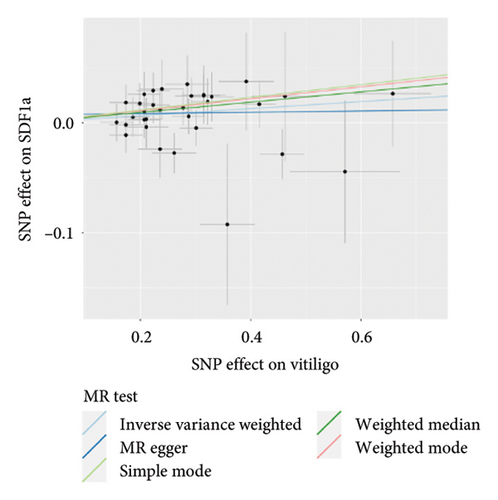
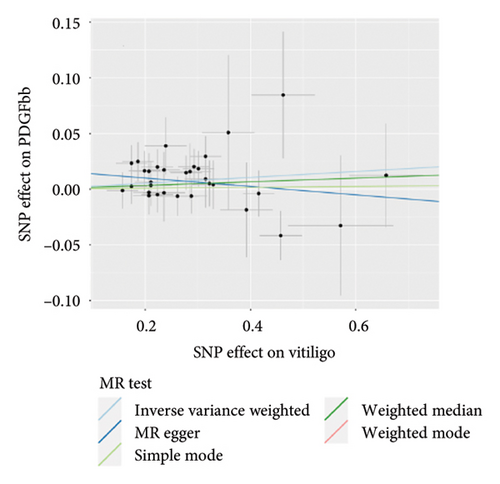
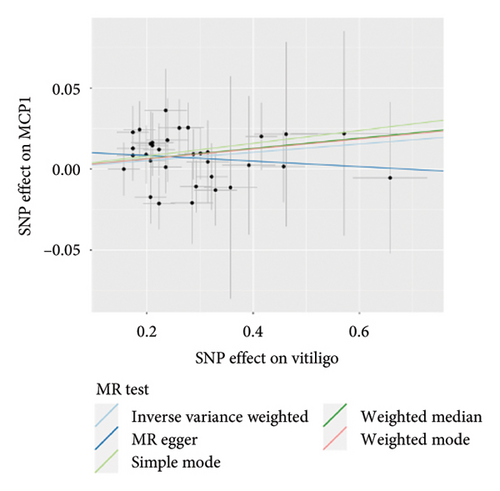
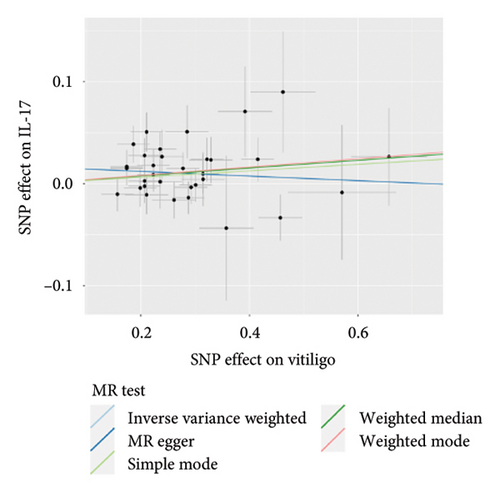
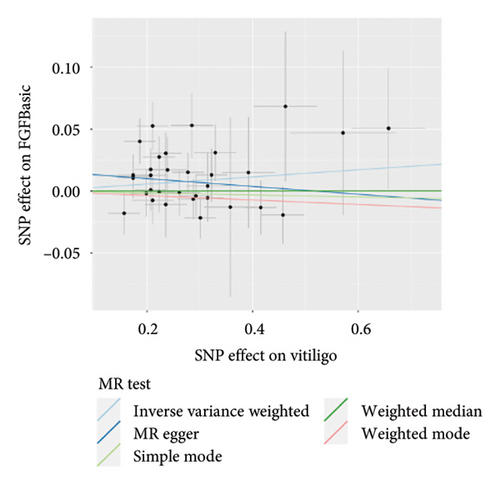
IVW analysis results were corrected by FDR, and we found SDF1a and IL-17 to be significant (p < 0.05). The weighted median analysis results for SDF1a (Beta: 0.047, 95% CI: 0.010–0.084, p = 0.013, and pfdr = 0.039) and IL-17 (Beta: 0.038, 95% CI: 0.001–0.075, p = 0.042, and pfdr = 0.106) were slightly different. The results obtained by the other four analysis methods were not significant (p > 0.05). The IVW analysis results of FGFBasic, MCP1, and PDGFbb were no longer significant after FDR correction (Table S5). The funnel plots in Figure 6 demonstrated the five positive cytokines before FDR correction, whereas Figure S2 and Figure S3 presented forest plots and leave-one-out sensitivity analyses.
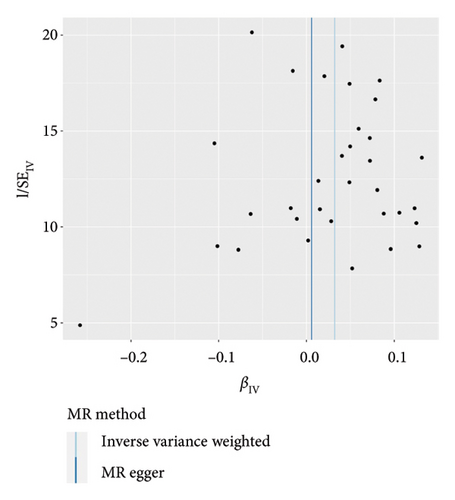
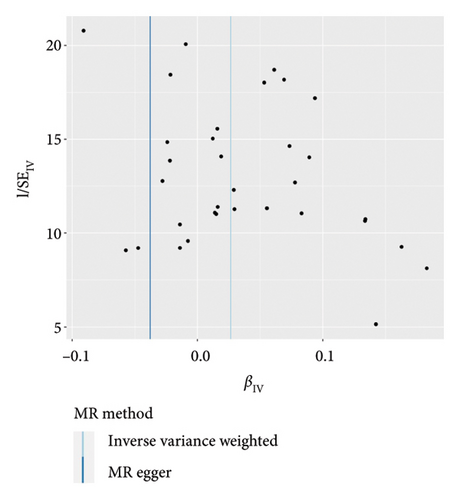
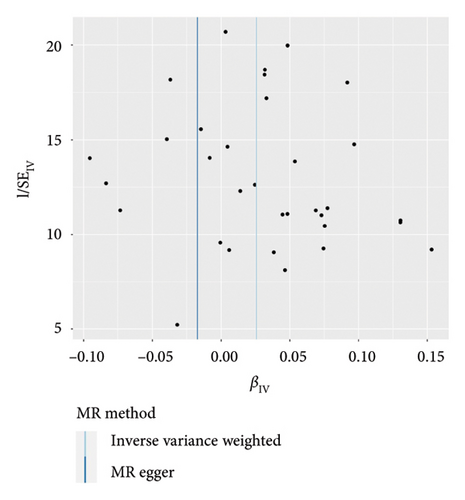
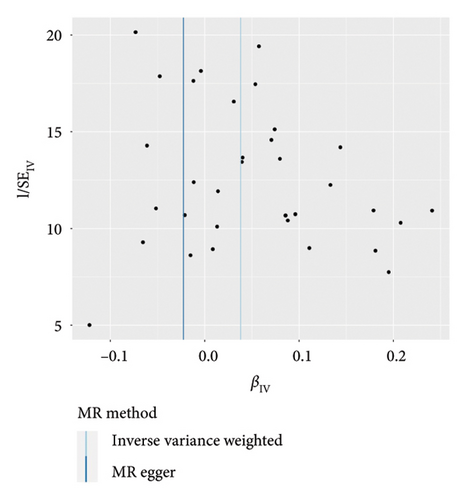
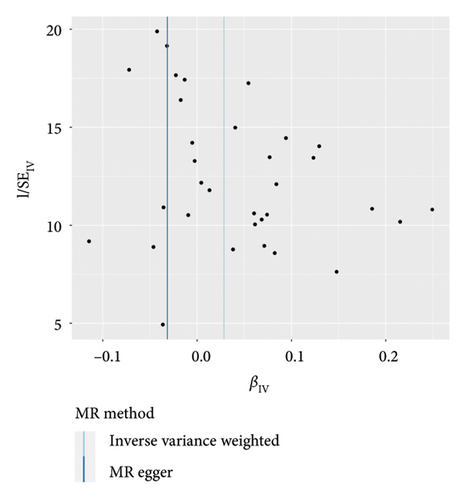
In the sensitivity test of this study, the results of MR-Egger intercept Cochrane’s Q analysis indicated that there were no pleiotropy and heterogeneity among IVs (p > 0.05). We also performed MR-PRESSO global test, which demonstrated that even after filtering out the SNPs (rs706779 and rs7137828), the corrected results remained consistent (p > 0.05) (Table S6). Consequently, the results obtained by our research team regarding the potential genetic predictive relationship between vitiligo and 41 inflammatory cytokines exhibit robustness.
4. Discussion
In this analysis involving two-sample MR, we investigated the genetic predictive connection between 41 biomarkers, specifically growth factors, interleukins, chemokines, and five additional cytokines, functioning as points of exposure, and vitiligo as the resulting outcome. Our findings suggested that genetic predictive IL-18 and IL-8 may be the upstream pathogenesis of vitiligo. The gene predicted that elevated IL-18 would increase the risk of vitiligo, but elevated IL-8 would decrease the risk of vitiligo. In addition, if vitiligo is defined as an exposure variable in MR, it may prompt an increase in SDF1a, PDGFbb, MCP1, IL-17, and FGFBasic levels. However, after FDR correction, the p values related to PDGFbb, MCP1, and FGFBasic were no longer statistically significant (pfdr > 0.05). In addition, there is no reverse genetic predictive relationship between certain cytokines and vitiligo. To summarize, genetic predictive IL-18 and IL-8 could potentially contribute to the development of vitiligo, whereas SDF1a, PDGFbb, MCP1, IL-17, and FGFBasic are more inclined to be involved in the progression of the disease.
Although the etiology of vitiligo is unknown, autoimmune theory has been proposed to be related to it, and now there is convincing evidence that cytokines play important roles in the pathogenesis of autoimmunity [19]. IL-18 is a powerful proinflammatory cytokine, which is involved in host defense against infection and regulates innate and acquired immune response. It has been reported that the level of IL-18 in blood of some patients with immune-related diseases is increased, and animal model tests clearly show that IL-18 is involved in the progress or development of some inflammatory and autoimmune diseases [20]. Inflammatory corpuscles, when assembled, trigger the activation of caspase-1, leading to the maturation and secretion of pro-inflammatory cytokines such as IL-1b and IL-18. This process also induces thermal death to eliminate infectious factors. The importance of inflammatory bodies in regulating immune response has been recognized with the discovery of gene polymorphism encoding inflammatory body components and their association with abnormal production of IL-1b and IL-18 in autoimmune and hereditary periodic fever syndrome [21]. NOD-like receptor family pyrin domain containing 1 (NLRP1) is an inflammatory body protein that plays a crucial regulatory role in the immune system. NLRP1 haplotype is associated with vitiligo susceptibility [22, 23]. A study found that SNP located in the promoter region and NLRP1 gene coding sequence was strongly and independently associated with vitiligo and vitiligo-related autoimmune diseases [24]. Thus, NLRP1 may affect the pathogenesis of vitiligo by mediating inflammatory factors such as IL-1b and IL-18. However, in a clinical controlled trial, it was found that there was no significant difference in serum IL-18 concentration between vitiligo patients and healthy subjects, and no correlation between IL-18 and vitiligo was found [25]. While IL-18 might contribute to the initial development of vitiligo, there is no substantial association observed with the subsequent progression of the disease. Therefore, the significant expression of IL-18 may not be detected after the onset of vitiligo. As predicted by the genetic level, these studies indicated that the elevated level of IL-18 may be related to the progression of some inflammatory and autoimmune diseases, thus affecting the pathogenesis of vitiligo.
IL-8 is one of the most widely studied leukocyte chemoattractants, which plays a role in attracting and activating leukocytes during inflammation. More and more evidences demonstrated the clinical relevance of IL-8 in autoimmune diseases [26]. Patients with active vitiligo showed increases in several pro-inflammatory (IL-6, IL-8, TNF-α, IFN-γ, and IL-1β) and some anti-inflammatory/immunomodulatory (IL-5 and IL-10) cytokines. The increased production of reactive oxygen species in red blood cells and neutrophils of patients with active vitiligo leads to lipid peroxidation and DNA damage. Furthermore, the decrease in antioxidant capacity creates an environment that promotes oxidation, tissue damage, and potentially new antigen production, thereby contributing to disease progression [27]. Moreover, research has indicated that the release of high mobility group box-1 protein from melanocytes fosters the secretion of CXCL16 and IL-8 from keratinocytes by binding to the receptor affiliated with advanced glycation end products. Subsequently, this binding activates both nuclear factor NF-kappaB and the extracellular signal-regulated kinase signaling pathway. Consequently, these mechanisms play a crucial role in the generation of autoimmunity due to oxidative stress in vitiligo [28]. Present models exploring the pathogenesis of vitiligo hypothesize that oxidative stress triggers cellular deterioration, which includes disruptions in protein maturation within the endoplasmic reticulum, induction of the unfolded protein response (UPR), and the expression of UPR-regulated chemokines such as IL-6 and IL-8. These chemokines exhibit the ability to recruit immune constituents to the skin, ultimately resulting in the targeting and destruction of melanocytes and the subsequent promotion of autoimmune responses throughout the course of the disease [29]. These studies indicated that IL-8 tends to be overexpressed in vitiligo patients, but our gene prediction in this study suggested that the increase of IL-8 can reduce the risk of vitiligo. This contradiction may be due to the limited investigation of IL-8 levels before the onset of vitiligo in patients. Whether IL-8 can be a tool for us to predict inflammatory factors of vitiligo is still worth exploring. In addition, there may be interaction between IL-18 and IL-8. While only limited research has addressed this issue, the investigation into the involvement of these two cytokines in initiating immune responses and the subsequent cascade of events within the context of diseases remains an important area requiring resolution.
IL-17, which is a group of six cytokines consisting of IL-17A–IL-17F [30], has been extensively researched for its role in immune regulation. Among these cytokines, IL-17A (also known as IL-17) and IL-17F demonstrate the highest similarity [31]. Previous studies have shown that IL-17 is involved in the pathogenesis of various autoimmune diseases and is closely related to vitiligo, which supports our findings [32, 33]. But the mechanism is still unclear. Studies have indicated that vitiligo patients exhibit increased levels of IL-17 at the systemic, tissue, and cellular levels [6, 9, 34–36]. Animal experiments have also confirmed an increase in IL-17 levels in mice [37–39]. There is a vital aspect of vitiligo improvement that involves various treatment methods, including ultraviolet B phototherapy, which has the capacity to regulate IL-17 levels [40, 41]. The correlation between the mentioned levels and the activity as well as the severity of the disease is highly significant, demonstrating its potential influence on the progression and assessment of severity. Exogenous IL-17 directly antagonizes factors related to melanocyte function and survival, which may lead to clinical depigmentation [42]. IL-17 induces cellular stress microenvironment in melanocytes and promotes autophagic cell apoptosis in vitiligo [43].
The pathogenesis and development of vitiligo are closely related to the immune status of the body, mainly achieved through immune cell surface molecules, cytokines, and inflammatory mediators. Abnormal levels of related cytokines may also contribute to changes in the condition. However, using cytokines as conventional biomarkers may not be suitable due to their low expression levels, which makes them difficult to measure accurately. In addition, the interpretation of results is limited by various methodological variables, such as different definitions of disease activity, varying sample sizes and patient characteristics, and diverse laboratory techniques. Consequently, there is a relatively high incidence of nonconfirmatory or even conflicting results. Sensitivity and specificity analysis have not yet been conducted for most circulating biomarkers. Meanwhile, a good biomarker needs to be clearly separated between stable and active patients in order to achieve sufficient stratification. Over the last few years, significant active indicators have been identified in the skin as well as the bloodstream. To delve deeper into the involvement of indicators in the development of vitiligo, it is imperative to conduct extensive multisite investigations, aiming to authenticate the application of these indicators in clinical experiments and everyday procedures. Currently, various treatment methods for vitiligo are not ideal. The underlying cause of unsatisfactory treatment outcomes remains unclear, but our investigation proposes that these cytokines may be subsequent outcomes of vitiligo progression. Therefore, treatments targeting these cytokines may not effectively hinder disease progression from the onset.
This study is the initial MR investigation to assess the genetic predictive relationship between 41 inflammatory cytokines and vitiligo. However, the limitations of this study should be taken into account. First, this study was conducted in a European population, and it is uncertain whether these findings apply to other populations. Therefore, future research on the genetic predictive relationship between inflammatory cytokines and vitiligo should include samples from different races to enhance the generalizability of the results. Second, the specific diagnostic criteria for vitiligo vary depending on the disease stage, classification, and severity. For instance, in 2011, an international consensus classified vitiligo into nonsegmental vitiligo (NSV) and segmental vitiligo (SV) due to their different prognostic significance [44]. Therefore, future research can explore these subgroups in more detail. Lastly, the limitations associated with MR analysis prevent the accurate examination of the second and third hypotheses, potentially leading to bias. Further research is necessary to validate our findings and explore their potential application in clinical diagnostics and treatment strategies.
5. Conclusion
This study is the first to reveal the genetic predictive association between inflammatory cytokines and vitiligo. The results suggested that IL-18 and IL-8 may be factors related to the etiology of vitiligo, while SDF1a, PDGFbb, MCP1, IL-17, and FGFBasic may be involved in the downstream development of vitiligo.
Ethics Statement
This study was solely based on publicly accessible deidentified data from published studies and summary database. No separate ethical approval was needed in this study.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding
No funding was received for this research.
Acknowledgments
Special thanks to the related investigators for sharing the GWAS summary statistics included in this study. All data used in the current study are publicly available GWAS summary data.
Supporting Information
Additional supporting information can be found online in the Supporting Information section.
Open Research
Data Availability Statement
Publicly available datasets were analyzed in this study. The data on inflammatory cytokines can be found here: https://doi.org/10.5523/bris.3g3i5smgghp0s2uvm1doflkx9x. The data on vitiligo are openly available in the GWAS Catalog (GCST004785) (https://www.ebi.ac.uk/gwas/studies/GCST004785).




