Topological Descriptors of Colorectal Cancer Drugs and Characterizing Physical Properties Via QSPR Analysis
Abstract
Topological descriptors and QSPR analysis are statistical techniques that are highly beneficial for analyzing various physical and chemical characteristics of molecular graphs without necessitating expensive and time-consuming laboratory experiments. The topological descriptor alters the compound to a number and helps in finding physicochemical properties. It more correctly reproduces the theoretical properties of drugs. In this article, the author investigated colorectal drugs capecitabine, leucovorin, tipiracil hydrochloride, etc. and implemented QSPR analysis. Physical qualities such as molar volume, complexity, polarity, and refractivity are the subject of the current study. The outcomes of this study allow for more effective physical property prediction through the use of QSPR models. First, we calculate Tds and secondly perform QSPR analysis. Current work on TIs and QSPR modeling shows a good correlation with physical properties. Moreover, estimated drug results depict and predict the physical properties in an efficient way.
1. Introduction
Colorectal cancer (CRC) is the third most frequent cancer and the leading cause of cancer death in the United Kingdom. Despite major improvements in our understanding of early detection and prevention, the disease ranks third in terms of cancer-related deaths [1]. In Asia, CRC is the fourth most common cancer-related cause of death, and its incidence is rising. CRC is one of the main causes of cancer fatalities in Europe [2] and Asia. Epidemiological data have revealed that several aspects with family history, excessive meat ingesting, alcohol use, and genetic changes impact the risk of colorectal neoplasia [3]. In CRC, cells in the colon proliferate out of control. The remaining waste material from the colon enters the rectum, the stomach’s last six inches of digestive tract. The depth to which a CRC penetrates the wall and if it has progressed outside the colon or rectum determine the stage of the disease. However, the exact mechanism underlying colorectal carcinogenesis is still not known. CRC may have both hereditary and environmental causes, unlike other complicated diseases [4]. With 783,000 new cases per year, CRC is a serious health issue that accounts for 9% of all cancers in the globe [5]. It peaked in the United States in 1994 with 131,000 new cases and 57,000 fatalities. There is growing evidence that CRC can spread to numerous organs and that metastatic illness is the leading cause of death in CRC patients. The liver is the CRC metastases’ preferred target [6–8]. The second most frequent disease in the world and the second largest cause of cancer death, sporadic CRC accounts for 75% of all CRC cases [9]. Adenoma is the lesion that occurs first in a series that could last 10–15 years before CRC. If men and women are taken into account jointly, CRC is the most prevalent malignancy in the world, with more than 1,200,000 new cases per year. North America, Europe, Australia, and New Zealand have the greatest incidence rates. Moreover, lung cancer is the second most common cause of cancer-related fatalities in these populations. Since premalignant polyps account for more than 85% of tumor emergence, CRC may be prevented. Therefore, the goal of CRC screening is to lower mortality by detecting people who have neoplastic lesions that are presymptomatic and who may need additional testing and treatment. Testing procedures should be affordable, regarded as appropriate, sensitive, focused, and secure. However, none of the suggested assays for CRC screening that are currently available meet these conditions [10]. A CRC-related death occurs every 9 minutes, according to the reports. In addition, it ranks third among the most prevalent malignancies in the WHO EMRO region, behind lung and breast cancer in women and cervical cancer in males [11,12].
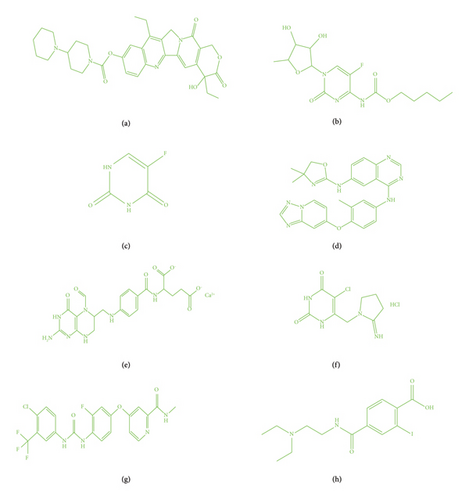
CRC slays a number of individuals. Researchers create and study novel medications. Their creation is a difficult job because of costly, time-consuming, and incredibly tough. Many drugs trial is imposed to stop this deadly disease, and numerous drug tests are imposed to combat lethal disease. It demands swift finding and medication that will device the disease. Eight drugs’ medicines irinotecan, capecitabine, fluorouracil, leucovorin, tipiracil hydrochloride, regorafenib, tucatinib, and bevacizumab are harmless and are more effective in nature for well-being community. Figure 1 displays the aforementioned drugs. All Tds are important and show a noteworthy role in chemical graphs as they can expose and predict hidden molecular graph properties. Tds not only has applications in science such as cheminformatics and bioinformatics but also has a substantial role in quantitative structure–property relationship (QSPR) [13–22]. To predict drug bioactivity, ABC, Wiener, and Randic index are suitable invariants. These topological indices serve an important role in current drug design by connecting molecular structure and biological activity. Their predictive capability speeds up drug discovery, improves knowledge of structure–activity correlations, and aids in the development of safer, more effective therapies. In this article, the author calculated Tds for CRC drugs. Correspondingly, CRC treatment molecular graphs are meticulously studied with Tds and imposed QSPR (modals). Calculated results on CRC drugs depict QSPR, and Tds have a good relation. Siddiqui et al. [23] made a contribution toward titanium that mathematical modeling gives us a proficient method for changing over an issue from an application region such as science, physical science, and science into a numerical system. Titanium exists in four kinds of oxides, and various applications exist in textures, papers, food varieties, and medication. Titanium dioxide is a synthetically steady and climate-well-disposed oxide that exists in unmistakable translucent stages such as rutile, brookite, anatase, baddeleyite, columbite, and fluorite. Research on novel drugs in the treatment of CRC was discussed by Havare [13] and gave suggestion that drug discovery needs huge costs to develop and intricate processes so are efficiently predicted with this technique. QSPR modeling of blood cancer drugs was investigated by Nasir et al. [14] and observed as a suitable model for it. Being efficient and wide spread, QSPR studies for various Tds for drug structures inspired to investigate CRC drugs. The objective is to thoroughly investigate in implementing Tds to probe properties and its QSPR analysis on CRC molecular graphs in healing management. Diabetes disease drugs were discussed by Parveen et al. [15]. They impose degree base Tds with the aid of regression analysis and crafted well-developed model for RA disease. Synthetic organizations of silicate and hexagon are very much portrayed by Kulli, Chaluvaraju, and Asha [16] and made a near investigation of the designs. The deep behavior of the networks can be better understood with the help of these findings. A computational base method is applied by Adnan et al. [17] for unequivocal degree and distance-based Tds for few organizations. HIV is a deadly sickness all through the world since it has no legitimate fix to date whereas medication preliminaries are finished to battle the illness, and a proper QSPR model is carried out by Farooq et al. [18]. The QSPR illustration of heart-related drugs is observed in [19], and Bondy and Murty [20] achieved curvilinear QSPR learning of blood medicines and demonstrated right model for it. Siddiqui et al. [21] track down multiplicative Zagreb indices and found useful results of some graphs. Molecular graphs of rheumatoid arthritis worked out with topological descriptors in [24]. Drugs for vitiligo where results are provided to beautifully illuminate the subject [25]. Khan et al. [26] discussed in depth QSPR study on bladder cancer drugs. The skin cancer halts all over the world. The investigation of drugs aims to thoroughly explore and create new medications in a highly effective manner [27]. Zaman et al. [28] computed valuable formulas, and results are effective in QSPR analysis. Wang et al. [29] mentioned cancer therapy. Each year, this sickness affects up to 10 million people worldwide. Researches on molecular graphs Tds and cancer-treating drugs via QSPR correlate the physical properties. The author’s deliberate study looks at some drugs that are efficiently employed in CRC therapy. The work on current study depends on Tds on many chemicals. In this scenario, the author studied degree-based topological descriptors on CRC drugs.
2. Materials and Methods
The molecular graphs of medications for CRC treatment drugs have been examined in this paper. Approaches such as edges partitioning, vertex partitioning, and computational methods are used to derive the topological indices of pharmaceuticals. Graph G(V, E) is simple and connected. The V(G) and E(G) represent vertex and edge sets, respectively. du denotes the degree of vertex. We implement the following Tds by Shigehalli and Kanabur [30]:
Metastatic cancer of the colon which is treated with irinotecan is an antineoplastic enzyme inhibitor. This exhibits chemical formula C33H38N4O6. Antineoplastic is another enzyme inhibitor, and irinotecan is primarily used to treat CRC. In addition, it can be administered for adults with locally advanced rectal cancer prior to surgery. Capecitabine is recommended for treating cancer. It can also be used in combination with docetaxel when the illness has progressed after receiving anthracycline-containing chemotherapy in the past. Capecitabine is for the treatment of adult patients with unresectable or metastatic gastric, esophageal, or gastroesophageal junction. C4H3FN2O2 is its chemical composition. A pyrimidine compound exhibiting antimetabolite properties targeting tumors. By preventing thymidylate synthetase from converting deoxyuridylic acid to thymidylic acid, it prevents DNA synthesis. Leucovorin is a folate analog that is used to treat megaloblastic anemia and CRC. Despite this difference in activity, leucovorin and levoleucovorin are both used as folate analogs to prevent the harmful effects of folic acid antagonists such as methotrexate, which function by inhibiting the enzyme dihydrofolate reductase. They are suggested for use as rescue therapy after the administration of high doses of methotrexate in the treatment of osteosarcoma or for reducing the toxicity brought on by unintentional overdosage of folic acid antagonists. The use of tipiracil hydrochloride is recommended for the treatment of metastatic CRC that has already received treatment with chemotherapy which includes fluoropyrimidine, oxaliplatin, and irinotecan. Metastatic gastrointestinal stromal tumors, hepatocellular carcinoma, and metastatic CRC are all conditions that regorafenib is used to treat. C21H15ClF4N4O3 is the drug’s chemical formula. Multiple kinases are inhibited by the oral medication regorafenib. Advanced gastrointestinal tumors, hepatocellular carcinoma, and metastatic CRC are all treated with it. In April 2017, regorafenib received approval for the treatment of hepatocellular carcinoma. Previous research on tuberculosis, breast cancer, bladder cancer, and QSPR analysis of various Tds for various drugs instigated to work CRC. The author looks into the relation of molecular graph Tds and its QSPR modeling of CRC which is suggested in therapeutic management.
3. Quantitative Structure Analysis and Regression Models
QSPR analysis is a computational method for predicting a chemical compound’s properties or activities based on its molecular structure. The author used degree-based topological metrics and regression models to assess and forecast the efficacy of new medications for CRC illnesses. The structures of these drugs are given in Figure 1. The study of the QSPR revealed a strong connection between TIs and the physiochemical properties of drugs under investigation. In Tables 1 and 2, the author has tabulated calculations of the above Tds and physicochemical properties of molecular structures, respectively. This will show regression models of worked analysis.
| Drugs | SK | SK1 | SK2 | GO1 | GO2 | KCD1 | KCD2 |
|---|---|---|---|---|---|---|---|
| Irinotecan | 123 | 153 | 318.5 | 552 | 1636 | 394 | 782 |
| Capecitabine | 62 | 72 | 152.5 | 268 | 736 | 196 | 362 |
| Fluorouracil | 21 | 23 | 50 | 88 | 226 | 66 | 116 |
| Leucovorin | 86 | 99.5 | 210.5 | 371 | 1004 | 272 | 498 |
| Tipiracil hydrochloride | 41 | 47.5 | 101 | 177 | 482 | 130 | 240 |
| Regorafenib | 85 | 97.5 | 211 | 365 | 994 | 270 | 504 |
| Tucatinib | 101 | 119.5 | 252.5 | 441 | 1212 | 322 | 606 |
| Bevacizumab | 46 | 51.5 | 109.5 | 195 | 510 | 144 | 254 |
| Molar volume | ||||
|---|---|---|---|---|
| Regression model | Molecular descriptor | R2 | F | Sig |
| Logarithmic | SK (G) | 0.972 | 103.177 | 0.002 |
| SK1 (G) | 0.976 | 122.284 | 0.002 | |
| SK2 (G) | 0.976 | 122.094 | 0.002 | |
| GO1 (G) | 0.974 | 114.33 | 0.002 | |
| GO2 (G) | 0.969 | 93.57 | 0.002 | |
| M1 (G) | 0.972 | 103.177 | 0.002 | |
| M2 (G) | 0.976 | 122.284 | 0.002 | |
| KCD1 (G) | 0.973 | 107.11 | 0.002 | |
| KCD2 (G) | 0.978 | 135.197 | 0.001 | |
| ABC (G) | 0.965 | 83.395 | 0.003 | |
| RA (G) | 0.961 | 74.104 | 0.003 | |
| F (G) | 0.975 | 119.185 | 0.002 | |
| HM (G) | 0.976 | 122.094 | 0.002 | |
| Quadratic | SK (G) | 0.992 | 128.406 | 0.008 |
| SK1 (G) | 0.991 | 115.606 | 0.009 | |
| SK2 (G) | 0.993 | 146.931 | 0.007 | |
| GO1 (G) | 0.992 | 124.061 | 0.008 | |
| GO2 (G) | 0.991 | 116.181 | 0.009 | |
| M1 (G) | 0.992 | 128.406 | 0.008 | |
| M2 (G) | 0.991 | 115.606 | 0.009 | |
| KCD1 (G) | 0.992 | 126.676 | 0.008 | |
| KCD2 (G) | 0.994 | 155.744 | 0.006 | |
| ABC (G) | 0.994 | 156.128 | 0.006 | |
| RA (G) | 0.997 | 290.7 | 0.003 | |
| F (G) | 0.994 | 174.768 | 0.006 | |
| HM (G) | 0.993 | 146.931 | 0.007 | |
3.1. Topological Descriptors Calculation
Topological descriptors’ calculation of other CRC drugs follows the same process and given in Tables 1 and 3.
| Drugs | ABC | RA | GA | M1 | M2 | H | HM | F |
|---|---|---|---|---|---|---|---|---|
| Irinotecan | 34.57 | 20.82 | 47.78 | 246 | 306 | 20.20 | 1274 | 662 |
| Capecitabine | 18.80 | 11.85 | 24.96 | 124 | 144 | 11.27 | 610 | 322 |
| Fluorouracil | 6.65 | 4.20 | 8.52 | 42 | 46 | 3.93 | 200 | 108 |
| Leucovorin | 25.98 | 16.19 | 34.66 | 172 | 199 | 15.47 | 842 | 444 |
| Tipiracil hydrochloride | 12.34 | 7.58 | 16.30 | 82 | 95 | 7.20 | 404 | 214 |
| Regorafenib | 25.48 | 15.56 | 33.41 | 170 | 195 | 14.75 | 844 | 454 |
| Tucatinib | 29.26 | 17.42 | 39.87 | 202 | 239 | 16.90 | 1010 | 532 |
| Bevacizumab | 14.46 | 9.49 | 19.19 | 92 | 103 | 9.03 | 438 | 232 |
3.2. Curvilinear Regression Models
- (i)
(exponential equation)
- (ii)
Z = a ln(t1) (logarithmic equation)
- (iii)
(quadratic equation)
- (iv)
(cubic equation)
| Polarity | ||||
|---|---|---|---|---|
| Regression model | Molecular descriptor | R2 | F | Sig |
| Cubic | SK (G) | 0.999 | 1149.679 | 0.000 |
| SK1 (G) | 0.999 | 1308.204 | 0.000 | |
| SK2 (G) | 0.999 | 845.706 | 0.000 | |
| GO1 (G) | 0.999 | 1260.645 | 0.000 | |
| GO2 (G) | 0.999 | 871.324 | 0.000 | |
| M1 (G) | 0.999 | 1149.679 | 0.000 | |
| M2 (G) | 0.999 | 1308.204 | 0.000 | |
| KCD1 (G) | 0.999 | 1159.723 | 0.000 | |
| KCD2 (G) | 0.999 | 684.923 | 0.000 | |
| ABC (G) | 0.999 | 683.200 | 0.000 | |
| RA (G) | 0.995 | 202.808 | 0.000 | |
| F (G) | 0.998 | 493.268 | 0.000 | |
| HM (G) | 0.999 | 845.706 | 0.000 | |
| Exponential | SK (G) | 0.914 | 53.243 | 0.001 |
| SK1 (G) | 0.888 | 39.521 | 0.001 | |
| SK2 (G) | 0.896 | 43.225 | 0.001 | |
| GO1 (G) | 0.900 | 45.177 | 0.001 | |
| GO2 (G) | 0.919 | 58.866 | 0.001 | |
| M1 (G) | 0.914 | 53.243 | 0.001 | |
| M2 (G) | 0.888 | 39.521 | 0.001 | |
| KCD1 (G) | 0.910 | 50.766 | 0.001 | |
| KCD2 (G) | 0.883 | 37.917 | 0.002 | |
| ABC (G) | 0.935 | 71.701 | 0.000 | |
| RA (G) | 0.941 | 50.321 | 0.000 | |
| F (G) | 0.903 | 46.633 | 0.001 | |
| HM (G) | 0.896 | 43.225 | 0.001 | |
| Molar volume | ||||
|---|---|---|---|---|
| Regression model | Molecular descriptor | R2 | F | Sig |
| Cubic | SK (G) | 0.994 | 54.83 | 0.099 |
| SK1 (G) | 0.993 | 44.569 | 0.11 | |
| SK2 (G) | 0.995 | 61.453 | 0.093 | |
| GO1 (G) | 0.993 | 49.408 | 0.104 | |
| GO2 (G) | 0.992 | 43.669 | 0.111 | |
| M1 (G) | 0.994 | 54.83 | 0.099 | |
| M2 (G) | 0.993 | 44.569 | 0.11 | |
| KCD1 (G) | 0.994 | 55.175 | 0.099 | |
| KCD2 (G) | 0.995 | 64.835 | 0.091 | |
| ABC (G) | 0.995 | 68.533 | 0.089 | |
| RA (G) | 0.997 | 101.592 | 0.073 | |
| F (G) | 0.996 | 85.03 | 0.08 | |
| HM (G) | 0.995 | 61.453 | 0.093 | |
| Exponential | SK (G) | 0.911 | 30.621 | 0.012 |
| SK1 (G) | 0.881 | 22.218 | 0.018 | |
| SK2 (G) | 0.893 | 25.15 | 0.015 | |
| GO1 (G) | 0.895 | 25.577 | 0.015 | |
| GO2 (G) | 0.915 | 32.384 | 0.011 | |
| M1 (G) | 0.911 | 30.621 | 0.012 | |
| M2 (G) | 0.881 | 22.218 | 0.018 | |
| KCD1 (G) | 0.907 | 29.154 | 0.012 | |
| KCD2 (G) | 0.881 | 22.313 | 0.018 | |
| ABC (G) | 0.936 | 44.215 | 0.007 | |
| RA (G) | 0.949 | 56.038 | 0.005 | |
| F (G) | 0.904 | 28.143 | 0.013 | |
| HM (G) | 0.893 | 25.15 | 0.015 | |
| Molar refractivity | ||||
|---|---|---|---|---|
| Regression model | Molecular descriptor | R2 | F | Sig |
| Logarithmic | SK (G) | 0.959 | 115.999 | 0.000 |
| SK1 (G) | 0.963 | 131.414 | 0.000 | |
| SK2 (G) | 0.962 | 125.202 | 0.000 | |
| GO1 (G) | 0.962 | 125.218 | 0.000 | |
| GO2 (G) | 0.957 | 110.093 | 0.000 | |
| M1 (G) | 0.959 | 115.999 | 0.000 | |
| M2 (G) | 0.963 | 131.414 | 0.000 | |
| KCD1 (G) | 0.960 | 119.025 | 0.000 | |
| KCD2 (G) | 0.963 | 129.440 | 0.000 | |
| ABC (G) | 0.951 | 96.0680 | 0.000 | |
| RA (G) | 0.943 | 82.9300 | 0.000 | |
| F (G) | 0.959 | 118.437 | 0.000 | |
| HM (G) | 0.962 | 125.202 | 0.000 | |
| Quadratic | SK (G) | 0.980 | 100.494 | 0.000 |
| SK1 (G) | 0.982 | 107.74 | 0.000 | |
| SK2 (G) | 0.980 | 98.069 | 0.000 | |
| GO1 (G) | 0.981 | 105.211 | 0.000 | |
| GO2 (G) | 0.981 | 101.711 | 0.000 | |
| M1 (G) | 0.980 | 100.494 | 0.000 | |
| M2 (G) | 0.982 | 107.74 | 0.000 | |
| KCD1 (G) | 0.981 | 100.627 | 0.000 | |
| KCD2 (G) | 0.979 | 95.52 | 0.000 | |
| ABC (G) | 0.979 | 91.774 | 0.000 | |
| RA (G) | 0.976 | 81.483 | 0.001 | |
| F (G) | 0.978 | 88.229 | 0.000 | |
| HM (G) | 0.980 | 98.069 | 0.000 | |
| Molar refractivity | ||||
|---|---|---|---|---|
| Regression model | Molecular descriptor | R2 | F | Sig |
| Cubic | SK (G) | 0.982 | 53.956 | 0.004 |
| SK1 (G) | 0.982 | 54.327 | 0.004 | |
| SK2 (G) | 0.981 | 50.373 | 0.005 | |
| GO1 (G) | 0.982 | 54.01 | 0.004 | |
| GO2 (G) | 0.982 | 54.016 | 0.004 | |
| M1 (G) | 0.982 | 53.956 | 0.004 | |
| M2 (G) | 0.982 | 54.327 | 0.004 | |
| KCD1 (G) | 0.982 | 53.62 | 0.004 | |
| KCD2 (G) | 0.980 | 48.449 | 0.005 | |
| ABC (G) | 0.982 | 55.083 | 0.004 | |
| RA (G) | 0.980 | 48.388 | 0.005 | |
| F (G) | 0.979 | 46.589 | 0.005 | |
| HM (G) | 0.981 | 50.373 | 0.005 | |
| Exponential | SK (G) | 0.862 | 31.229 | 0.003 |
| SK1 (G) | 0.837 | 25.606 | 0.004 | |
| SK2 (G) | 0.844 | 27.067 | 0.003 | |
| GO1 (G) | 0.849 | 28.058 | 0.003 | |
| GO2 (G) | 0.867 | 32.703 | 0.002 | |
| M1 (G) | 0.862 | 31.229 | 0.003 | |
| M2 (G) | 0.837 | 25.606 | 0.004 | |
| KCD1 (G) | 0.858 | 30.252 | 0.003 | |
| KCD2 (G) | 0.831 | 24.641 | 0.004 | |
| ABC (G) | 0.883 | 37.599 | 0.002 | |
| RA (G) | 0.889 | 40.231 | 0.001 | |
| F (G) | 0.850 | 28.309 | 0.003 | |
| HM (G) | 0.844 | 27.067 | 0.003 | |
| Complexity | ||||
|---|---|---|---|---|
| Regression model | Molecular descriptor | R2 | F | Sig |
| Logarithmic | SK (G) | 0.863 | 31.572 | 0.002 |
| SK1 (G) | 0.875 | 34.857 | 0.002 | |
| SK2 (G) | 0.869 | 33.203 | 0.002 | |
| GO1 (G) | 0.870 | 33.438 | 0.002 | |
| GO2 (G) | 0.864 | 31.863 | 0.002 | |
| M1 (G) | 0.863 | 31.572 | 0.002 | |
| M2 (G) | 0.875 | 34.857 | 0.002 | |
| KCD1 (G) | 0.864 | 31.780 | 0.002 | |
| KCD2 (G) | 0.872 | 34.068 | 0.002 | |
| ABC (G) | 0.854 | 29.235 | 0.003 | |
| RA (G) | 0.854 | 29.235 | 0.003 | |
| F (G) | 0.864 | 31.668 | 0.002 | |
| HM (G) | 0.869 | 33.203 | 0.002 | |
| Quadratic | SK (G) | 0.935 | 28.791 | 0.004 |
| SK1 (G) | 0.941 | 32.106 | 0.003 | |
| SK2 (G) | 0.936 | 29.074 | 0.004 | |
| GO1 (G) | 0.939 | 30.84 | 0.004 | |
| GO2 (G) | 0.942 | 32.207 | 0.003 | |
| M1 (G) | 0.935 | 28.791 | 0.004 | |
| M2 (G) | 0.941 | 32.106 | 0.003 | |
| KCD1 (G) | 0.934 | 28.24 | 0.004 | |
| KCD2 (G) | 0.935 | 28.812 | 0.004 | |
| ABC (G) | 0.936 | 29.27 | 0.004 | |
| RA (G) | 0.951 | 38.662 | 0.002 | |
| F (G) | 0.929 | 26.32 | 0.005 | |
| HM (G) | 0.936 | 29.074 | 0.004 | |
| Complexity | ||||
|---|---|---|---|---|
| Regression model | Molecular descriptor | R2 | F | Sig |
| Cubic | SK (G) | 0.947 | 18.036 | 0.020 |
| SK1 (G) | 0.950 | 18.836 | 0.019 | |
| SK2 (G) | 0.946 | 17.611 | 0.021 | |
| GO1 (G) | 0.949 | 18.571 | 0.019 | |
| GO2 (G) | 0.952 | 19.938 | 0.017 | |
| M1 (G) | 0.947 | 18.036 | 0.020 | |
| M2 (G) | 0.950 | 18.836 | 0.019 | |
| KCD1 (G) | 0.946 | 17.602 | 0.021 | |
| KCD2 (G) | 0.945 | 17.221 | 0.021 | |
| ABC (G) | 0.952 | 19.884 | 0.018 | |
| RA (G) | 0.966 | 28.247 | 0.011 | |
| F (G) | 0.943 | 16.470 | 0.023 | |
| HM (G) | 0.946 | 17.611 | 0.021 | |
| Exponential | SK (G) | 0.912 | 51.737 | 0.001 |
| SK1 (G) | 0.896 | 43.194 | 0.001 | |
| SK2 (G) | 0.899 | 44.391 | 0.001 | |
| GO1 (G) | 0.904 | 47.151 | 0.001 | |
| GO2 (G) | 0.92 | 57.557 | 0.001 | |
| M1 (G) | 0.912 | 51.737 | 0.001 | |
| M2 (G) | 0.896 | 43.194 | 0.001 | |
| KCD1 (G) | 0.908 | 49.487 | 0.001 | |
| KCD2 (G) | 0.889 | 39.987 | 0.001 | |
| ABC (G) | 0.928 | 64.675 | 0.000 | |
| RA (G) | 0.942 | 81.073 | 0.000 | |
| F (G) | 0.900 | 44.963 | 0.001 | |
| HM (G) | 0.899 | 44.391 | 0.001 | |
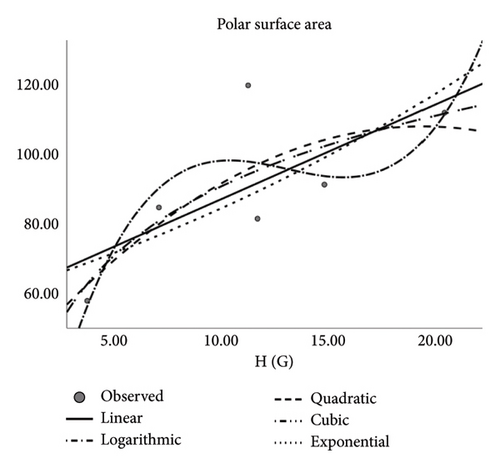
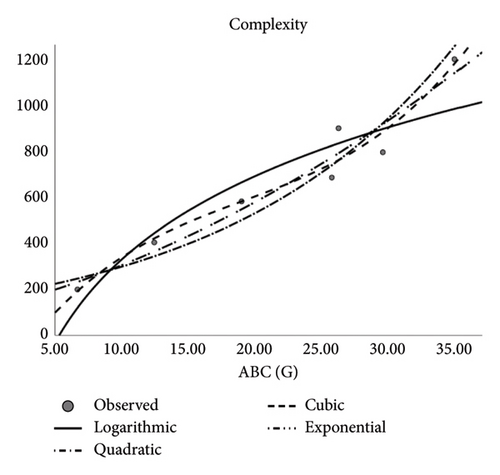
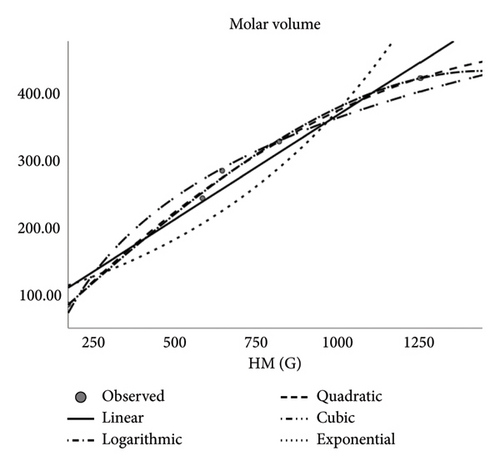

| Polarity | ||||
|---|---|---|---|---|
| Regression model | Molecular descriptor | R2 | F | Sig |
| Logarithmic | SK (G) | 0.956 | 107.600 | 0.000 |
| SK1 (G) | 0.959 | 118.303 | 0.000 | |
| SK2 (G) | 0.959 | 116.966 | 0.000 | |
| GO1 (G) | 0.958 | 114.361 | 0.000 | |
| GO2 (G) | 0.953 | 101.427 | 0.000 | |
| M1 (G) | 0.956 | 107.600 | 0.000 | |
| M2 (G) | 0.959 | 118.303 | 0.000 | |
| KCD1 (G) | 0.957 | 110.387 | 0.000 | |
| KCD2 (G) | 0.960 | 121.463 | 0.000 | |
| ABC (G) | 0.947 | 89.8870 | 0.000 | |
| RA (G) | 0.940 | 77.9960 | 0.000 | |
| F (G) | 0.958 | 114.203 | 0.000 | |
| HM (G) | 0.959 | 116.966 | 0.000 | |
| Quadratic | SK (G) | 0.999 | 2236.871 | 0.000 |
| SK1 (G) | 0.998 | 1152.827 | 0.000 | |
| SK2 (G) | 0.998 | 1224.954 | 0.000 | |
| GO1 (G) | 0.999 | 1637.881 | 0.000 | |
| GO2 (G) | 0.999 | 1659.673 | 0.000 | |
| M1 (G) | 0.999 | 2236.871 | 0.000 | |
| M2 (G) | 0.998 | 1152.827 | 0.000 | |
| KCD1 (G) | 0.999 | 2184.127 | 0.000 | |
| KCD2 (G) | 0.998 | 869.0050 | 0.000 | |
| ABC (G) | 0.998 | 1087.637 | 0.000 | |
| RA (G) | 0.995 | 376.6590 | 0.000 | |
| F (G) | 0.998 | 937.4390 | 0.000 | |
| HM (G) | 0.998 | 1224.954 | 0.000 | |
| Drugs | Polarity | Quadratic | Cubic | Logarithmic | Exponential |
|---|---|---|---|---|---|
| Predicted values with SK1 (G) index | |||||
| Irinotecan | 63.1 | 62.221 | 63.666 | 80.089 | 59.09 |
| Capecitabine | 32.6 | 36.558 | 37.829 | 30.337 | 39.033 |
| Fluorouracil | 10.2 | 13.201 | 11.023 | 16.863 | 8.666 |
| Leucovorin | 46.33 | 47.08 | 46.058 | 42.18 | 47.641 |
| Tipiracil hydrochloride | 25.618 | 27.237 | 22.618 | 27.965 | |
| Regorafenib | 44.8 | 46.378 | 45.519 | 41.181 | 47.10 |
| Tucatinib | 53.6 | 53.564 | 51.51 | 53.605 | 52.514 |
| Bevacizumab | 27.505 | 29.285 | 23.728 | 30.116 | |
| Predicted values with GO1 (G) index | |||||
| Irinotecan | 63.1 | 62.013 | 63.716 | 58.698 | 78.709 |
| Capecitabine | 32.6 | 36.405 | 37.725 | 38.988 | 30.328 |
| Fluorouracil | 10.2 | 13.191 | 10.841 | 8.608 | 16.570 |
| Leucovorin | 46.33 | 47.251 | 46.013 | 47.859 | 42.860 |
| Tipiracil hydrochloride | 25.346 | 27.137 | 27.671 | 22.342 | |
| Regorafenib | 44.8 | 46.668 | 45.566 | 47.414 | 42.005 |
| Tucatinib | 53.6 | 53.610 | 51.503 | 52.574 | 54.218 |
| Bevacizumab | 27.643 | 29.626 | 30.313 | 23.734 | |
| Predicted values with F (G) index | |||||
| Irinotecan | 63.1 | 61.847 | 63.772 | 58.517 | 77.724 |
| Capecitabine | 32.6 | 36.150 | 37.662 | 38.815 | 30.178 |
| Fluorouracil | 10.2 | 13.585 | 10.949 | 8.951 | 16.637 |
| Leucovorin | 46.33 | 46.804 | 45.556 | 47.597 | 42.375 |
| Tipiracil hydrochloride | 25.379 | 27.342 | 27.645 | 22.345 | |
| Regorafenib | 44.8 | 47.606 | 46.160 | 48.206 | 43.571 |
| Tucatinib | 53.6 | 53.493 | 51.259 | 52.541 | 54.132 |
| Bevacizumab | 27.262 | 29.425 | 29.853 | 23.492 | |
| Drugs | Molar volume | Quadratic | Cubic | Logarithmic | Exponential |
|---|---|---|---|---|---|
| Predicted values with KCD2 (G) index | |||||
| Irinotecan | 416.800 | 395.153 | 423.869 | 388.144 | 477.587 |
| Capecitabine | 240.500 | 269.777 | 297.330 | 280.361 | 235.334 |
| Fluorouracil | 84.600 | 147.013 | 101.597 | 121.102 | 155.472 |
| Leucovorin | 322.004 | 304.456 | 324.995 | 295.946 | |
| Tipiracil hydrochloride | 213.449 | 246.233 | 222.845 | 191.602 | |
| Regorafenib | 323.700 | 324.052 | 304.513 | 326.671 | 298.954 |
| Tucatinib | 339.000 | 355.544 | 313.556 | 352.463 | 355.018 |
| Bevacizumab | 220.368 | 255.807 | 230.779 | 196.176 | |
| Predicted values with RA (G) index | |||||
| Irinotecan | 416.800 | 381.524 | 425.656 | 374.801 | 447.450 |
| Capecitabine | 240.500 | 275.767 | 290.305 | 284.386 | 241.740 |
| Fluorouracil | 84.600 | 138.060 | 96.606 | 117.982 | 142.988 |
| Leucovorin | 334.445 | 300.409 | 334.450 | 325.630 | |
| Tipiracil hydrochloride | 204.297 | 244.809 | 212.704 | 180.326 | |
| Regorafenib | 323.700 | 326.801 | 295.958 | 328.083 | 311.848 |
| Tucatinib | 339.000 | 348.515 | 315.605 | 346.198 | 354.316 |
| Bevacizumab | 237.950 | 278.012 | 248.756 | 205.587 | |
| Predicted values with ABC (G) index | |||||
| Irinotecan | 416.800 | 381.988 | 425.618 | 376.648 | 448.140 |
| Capecitabine | 240.500 | 273.160 | 291.625 | 281.771 | 238.252 |
| Fluorouracil | 84.600 | 140.699 | 96.585 | 119.901 | 146.434 |
| Leucovorin | 331.548 | 298.029 | 332.154 | 317.657 | |
| Tipiracil hydrochloride | 208.000 | 246.736 | 216.195 | 183.925 | |
| Regorafenib | 323.700 | 327.960 | 296.441 | 329.128 | 311.358 |
| Tucatinib | 339.000 | 353.304 | 320.434 | 350.673 | 362.265 |
| Bevacizumab | 230.702 | 271.892 | 240.889 | 200.229 | |
4. Results and Discussion
5. Conclusions
The author observed correlation coefficients between the Td and some physical features of drugs applied to treat CRC that depicts how fine the said descriptors serve as interpreters. Descriptors (Tds) and QSPR analysis especially in the context of pharmaceutical and medical applications forecast the physical properties. Notably, molar refractivity, molar volume, polarity, and complexity are reliable and indicators for these predictions. However, the estimation of boiling point and polar surface area is less dependable. The medications used to treat CRC are the subject of this study. Pharmaceutical industry researchers and chemists may find use in the presented measurements provided in this work. Designing novel medications may benefit from these recent findings. Based on the discovered correlations, scientists can use the correlation coefficients of various medications to find the right composition for creating new drugs for emerging disorders. Multiple topological descriptors can be incorporated in unique ways to address inequalities by using a unified methodology. This article’s future recommendation suggests employing diverse forms to calculate the index’s extreme values.
Conflicts of Interest
The author declares no conflicts of interest.
Funding
The author does not have any funding available for this research.
Open Research
Data Availability Statement
All the data are available inside the manuscript, and there are no hidden data.




