Increased Long Noncoding RNA HCG18 Indicates the Adverse Prognosis and Deteriorating Development of Diffuse Large B Cell Lymphoma Through Suppressing miR-494-3p
Abstract
Objectives: Diffuse large B cell lymphoma (DLBCL) is a common and malignant subtype of lymphoma. Dysregulated lncRNA HCG18 has been reported to serve as tumor regulators in various human cancers. The expression and significance of lncRNA human leukocyte antigen complex Group 18 (HCG18) in the occurrence and development of HCG18 were confirmed in this study aiming to provide a potential biomarker for the screening and monitoring of DLBCL.
Methods: This study enrolled 82 DLBCL patients and the HCG18 levels in tissues were detected using polymerase chain reaction (PCR). Kaplan–Meier and Cox analyses were employed to assess its prognostic significance. In DLBCL cells, HCG18 was silenced by cell transfection, and its function in cell growth and metastasis was evaluated by cell counting kit-8 (CCK8) and Transwell assays.
Results: HCG18 was significantly upregulated in DLBCL tumor tissues, predicting the adverse outcomes of DLBCL patients and being correlated with the advanced malignant subtypes, Ann-Arbor stage, and high international prognostic index (IPI) of patients. In vitro, HCG18 negatively regulated the expression of miR-494-3p in DLBCL cells. Silencing HCG18 dramatically suppressed the proliferation, migration, and invasion of DLBCL cells, which was reversed by the knockdown of miR-494-3p.
Conclusions: Upregulated HCG18 served as a prognostic biomarker of DLBCL and promoted the development of DLBCL by negatively modulating miR-494-3p, which provides a candidate target for the clinical therapy of DLBCL.
1. Introduction
Diffuse large B cell lymphoma (DLBCL) is a common subtype of lymphoma sourced from mature B cells, which is aggressive with significant heterogeneity [1]. In the past decades, the incidence of DLBCL increased rapidly and its mortality also accounted for a large percentage of cancer-related deaths. Due to the heterogeneity of DLBCL, there were large individual differences in the clinical symptoms [2, 3]. Additionally, the outcomes of DLBCL patients are also associated with various factors, such as age, clinical stage, international postintervention index, and the level of lactate dehydrogenase. The prevention and therapy strategies of DLBCL have been significantly improved in the past decades; however, drug resistance, recurrence, and other risk factors resulted in an unsatisfying overall survival rate [4]. There was a lack of unified criteria for DLBCL detection and monitoring, and numerous studies have focused on the molecular typing, screening, and prognosis of DLBCL, aiming to provide a reliable reference for the clinical management of DLBCL. Identification of sensitive and accurate biomarkers is of great urgency and has become a current research hot point.
Gene abnormal expression has been considered one of the major initiation factors of DLBCL occurrence and progression. The epigenetic changes, such as methylation of DNA, the modification of histone, and the regulation of noncoding RNAs (ncRNAs) have been evidenced to mediate disease development and have attracted huge attention in recent years [5, 6]. Long ncRNAs (lncRNAs) are critical members of ncRNAs, which regulate gene expression at epigenetic, transcriptional, and post-transcriptional levels, including transcriptional inhibition, competitive endogenous RNA, chromatin modification, and other regulations [7]. Some ultra-conserved elements exist in lncRNAs, which are always distributed in chromosome-fragile and tumor-related genomic regions. Therefore, the abnormal expression of lncRNAs is considered to be related to the onset and development of malignant tumors [8, 9]. For example, dysregulated lncRNA IRF2-3 and lncRNA ZNF667-AS1 were revealed to show high sensitivity and specificity in screening B-chronic lymphocytic leukemia and predicting the prognosis of patients [10]. lncRNA HCG18 has been demonstrated to regulate the progression of human cancers, such as clear cell renal cell carcinoma, osteosarcoma, and lung adenocarcinoma [11–13]. In a previous transcriptome study, several dysregulated mRNAs and ncRNAs were identified to be associated with the relapse of Hodgkin lymphoma, another major subtype of lymphoma. The potential of HCG18 to mediate the progression of Hodgkin lymphoma was noticed in this study, but there was a lack of confirmation by clinical data, and the regulatory mechanism has not been revealed [14]. Although DLBCL is a kind of non-Hodgkin lymphoma, HCG18 is also considered to play a role in the development of DLBCL.
According to the prevalent competing endogenous RNA (ceRNA) theory, lncRNAs were accepted to sponge miRNAs and further regulate tumor progression and other disease development. With the help of the lncRNA SNP v3 database, the ceRNAs of HCG18 were predicted, among which miR-494-3p was previously reported to mediate the regulation of DLBCL progression by lncRNA SBF2-AS1 [15]. Hence, its involvement in the function of HCG18 in DLBCL was assessed in the present study.
This study evaluated the significance of HCG18 in predicting the severity and prognosis of patients with DLBCL via clinical samples and data. The regulatory effect and mechanism of HCG18 on DLBCL cellular processes were also investigated with in vitro experiments, aiming to identify a novel biomarker and potential therapeutic target for DLBCL.
2. Materials and Methods
2.1. Study Subjects
There were 82 DLBCL patients enrolled in this study from January 2017 to December 2019 at People’s Hospital of Quzhou. The enrolled patients were composed of 47 males and 35 females with an average age of 54.76 ± 8.72 years. The clinicopathological features of patients are summarized in Table 1. This study had been approved by the Ethics Committee of People’s Hospital of Quzhou and informed consent had been obtained from all participants. DLBCL patients were primarily diagnosed and had never received any antitumor therapy. Patients with other malignant tumors or complications were excluded. All clinical records of enrolled patients were completed.
| Cases (no. 82) | Low-HCG18 (n = 39) | High-HCG18 (n = 43) | p | |
|---|---|---|---|---|
| Age | 0.666 | |||
| < 55 | 40 | 20 | 20 | |
| ≥ 55 | 42 | 19 | 23 | |
| Gender | 0.545 | |||
| Male | 47 | 21 | 26 | |
| Female | 35 | 18 | 17 | |
| Tumor size | 0.173 | |||
| < 3 | 44 | 24 | 20 | |
| ≥ 3 | 38 | 15 | 23 | |
| Ann-Arbor stage | 0.013∗ | |||
| I-II | 54 | 31 | 23 | |
| III-IV | 28 | 8 | 20 | |
| LDH | 0.086 | |||
| < 300 | 34 | 20 | 14 | |
| ≥ 300 | 48 | 19 | 29 | |
| IPI | 0.026∗ | |||
| < 3 | 24 | 16 | 8 | |
| ≥ 3 | 58 | 23 | 35 | |
| Extranodal sites | 0.404 | |||
| < 2 | 64 | 32 | 32 | |
| ≥ 2 | 18 | 7 | 11 |
- Abbreviations: IPI, international prognostic index; LDH, lactate dehydrogenase.
- ∗p < 0.05.
2.2. Sample Collection
Tumor and adjacent paracancerous tissues were collected from all participants during their surgery. Paracancerous tissues were collected 5 cm from the edge of the tumor tissues. Samples were frozen with liquid nitrogen and stored at −80°C for the following analyses.
2.3. Follow-Up Survey
The enrolled patients were followed up for 3–60 months via telephone or outpatient reviews to obtain the recovery conditions of patients. The endpoints were defined as recurrence, DLBCL-related development, and all-cause deaths.
2.4. Cell Culture
DLBCL cells (OCI-ly1, DB, SU-DHL-4, and Farage) and normal B cells were purchased from the Cell Bank of Type Culture Collection (China). Cells were cultivated in 10% FBS-containing RPMI-1640 culture medium at 37°C and were used at passage less than 10 times. Cells were available for the following experiments when the fusion reached 70%–80%.
2.5. Cell Transfection
Available cells (5 × 105 cells/well) were seeded into 6-well plates and supplied with a completed culture medium. Cells were incubated with siRNA-HCG18 (si-HCG18, 10 nM) or negative control (si-NC, 10 nM) using Lipofectamine 2000 (Invitrogen, USA). The mixture was centrifugated and resuspended in a fresh culture medium after 24 of incubation. The transfection efficiency was evaluated when the cell confluence was over 90% about 48 h of cell transfection.
2.6. Real-Time Quantitative PCR
Total RNA was isolated from cells (1 × 105 cells/well) or tissue samples using TRIzol reagent (Invitrogen, USA). Isolated RNA was reverse transcribed into cDNA using a ReverTra Ace qPCR RT Kit (for HCG18, TOYOBO, Japan) and a Mir-X miRNA First-Strand Synthesis kit (for miR-494-3p, Clontech Laboratories, USA) amplified on the LightCycler 96 PCR system (Roche, Switzerland) with SYBR-Green kit (TOYOBO, Japan). The experimental primers are as follows: forward: HCG18 5′-ATCCTGCCAATAGATGCTGCTCAC-3, HCG18 reverse: 5′-AGCCACCTTGGTCTCCAGTCTC-3′; miR-494-3p forward, 5′-GGGTGAAACACACACGGGAA-3′, miR-494-3p reverse, 5′-GGCAGGTCCGAGGT-3′. The relative expression of HCG18 and miR-494-3p was calculated with the 2−ΔΔCT method normalized to 18s RNA (forward 5′-CGGACATCTAAGGGCATCACA-3′, reverse 5′-AACGAACGAGACTCTGGCATG-3′) and cel-miR-39 (forward 5′-CGTATGAGCG TCACCGGGTGTAAATCA-3′), respectively.
2.7. Dual-Luciferase Reporter Assay
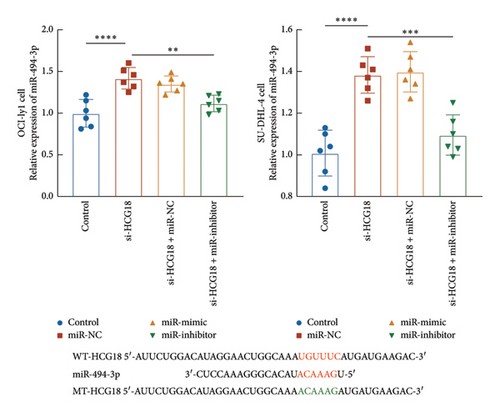
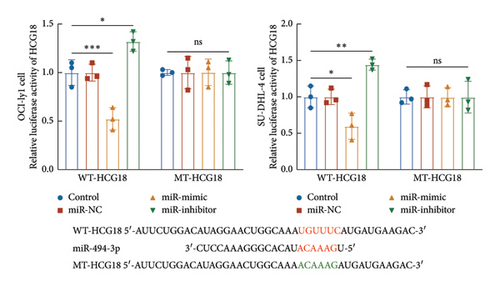
miR-494-3p was predicted as a ceRNA of HCG18 with several binding sites from the public lncRNASNP database (version 3.0, https://gong_lab.hzau.edu.cn/lncRNASNP3/#!/). The binding sites were cloned into the pGL3 vector (Promega, USA) to establish the wild-type plasmid, while the mutant-type plasmid was constructed by cloning the mutant sites (Figure 1(b)). DLBCL cells (1 × 105 cells/well) were co-transfected with miR-494-3p mimic, inhibitor, or negative controls and established plasmids using Lipofectamine 2000 at room temperature. The relative luciferase activity of HCG18 was detected by the Dual-luciferase reporter system (Promega, USA) with Renila as the internal reference.
2.8. Cell Counting Kit-8 (CCK8) Assay
Available cells were seeded into 96-well plates (1 × 105 cells/well) with triple repetitions of each well. A total of 90 μL completed culture medium was added to each well and the plates were incubated for 24, 48, 72, and 96 h. Then, CCK8 reagent (Beyotime, China) and gently tap the plates to blend the mixture. The plates were analyzed with a microplate reader after incubating at 37°C for 2 h. The absorbance at 450 nm was used to evaluate cell proliferation.
2.9. Transwell Assay
Available cells (1 × 105 cells/well) were seeded into the upper layers of 24-well Transwell chambers and supplied with an FBS-free culture medium. The bottom chamber was filled with the completed culture medium. For the invasion assessment, the Matrigel was diluted with an FBS-free culture medium and pre-coated in the upper chamber before seeding cells. The Transwell chambers were incubated at 37°C for 24–48 h and then the cells on the subsurface of the upper chamber were soaked with 4% polyformaldehyde for 15 min. After washing with PBS three times, cells were further stained with crystal violet and counted under an optical microscope.
2.10. Employment of Public Databases
The downstream targets of miR-494-3p were predicted from miRDB (https://mirdb.org/), Target Scan (https://www.targetscan.org/vert_80/), miRWalk (https://mirwalk.umm.uni-heidelberg.de/), and STARBASE (https://mirwalk.umm.uni-heidelberg.de/). The function of the enriched genes was annotated from the Database for Annotation, Visualization and Integrated Discovery (DAVID) database (https://david.ncifcrf.gov/).
2.11. Statistical Analyses
Experimental data were analyzed and plotted with GraphPad Prism 9.0. Clinical analyses were conducted with SPSS 26.0 software. Specifically, difference comparison was conducted with a Student’s t-test or one-way ANOVA (p < 0.05). The chi-square test was employed to evaluate the association of HCG18 with patients’ clinical features. The follow-up data were analyzed by Kaplan–Meier and multivariate Cox regression analyses.
3. Results
3.1. Expression and Significance of HCG18 in DLBCL
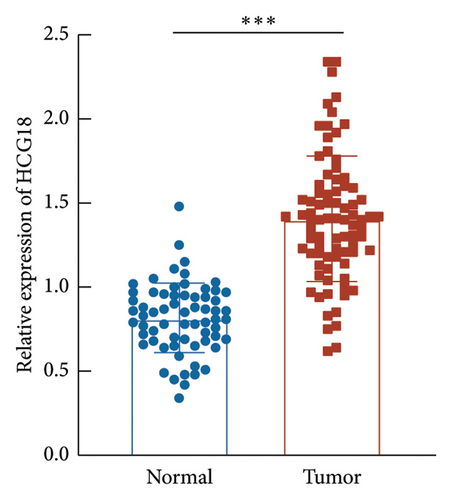
HCG18 in DLBCL tumor tissues was significantly higher than in normal tissues (Figure 2(a)). According to the mean value of HCG18 expression in tumor tissues (1.41), a low-HCG18 group and a high-HCG18 group were established. In the low-HCG18 group, DLBCL patients showed a better 5-year development-free survival rate than the high-HCG18 group (Figure 2(b)). HCG18 was also revealed to possess prognostic significance serving as an independent indicator (hazard ratio, HR = 9.104, 95% confident interval, 95% CI = 1.921–43.139) together with tumor size (HR = 3.455, 95% CI = 1.365–8.745), Ann-Arbor stage (HR = 4.595, 95% = 1.782–11.847), LDH level (HR = 3.193, 95% CI = 1.221–8.350), IPI scores (HR = 7.930, 95% CI = 1.500–19.148), and subtypes (HR = 4.445, 95% CI = 1.207–16.363, Figure 2(c)).
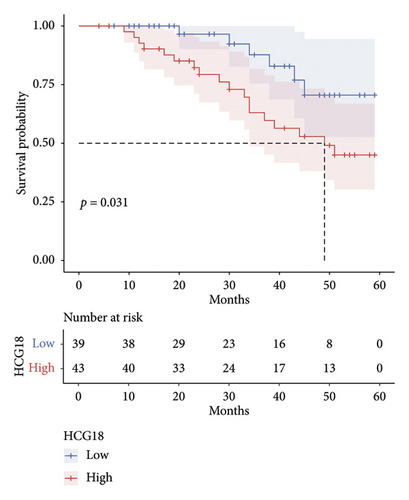
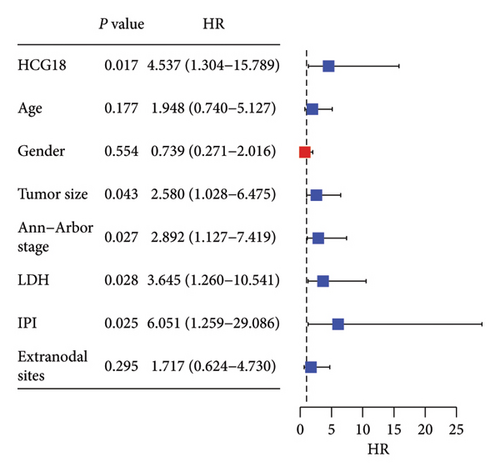
The high HCG18 level in DLBCL patients was associated with the malignant subtype (p < 0.001), advanced Ann-Arbor stage (p = 0.013), and high IPI scores (p = 0.026), which represented the malignant and severe development of DLBCL (Table 1).
3.2. The Function of HCG18 in DLBCL Cells
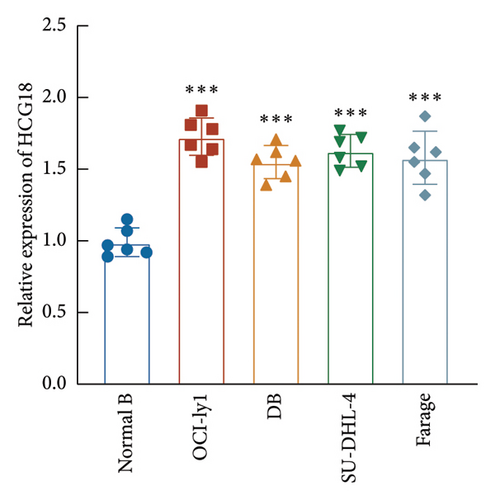
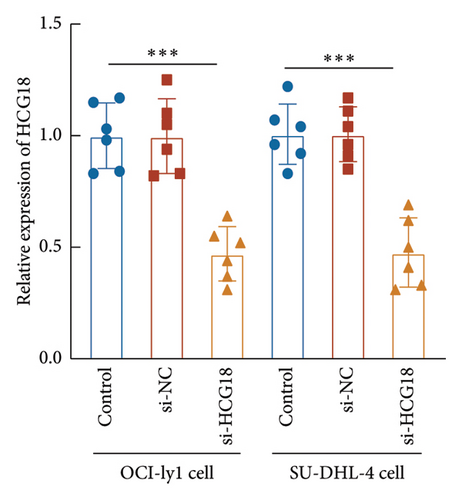
Consistently, a significant upregulation of HCG18 was also observed in the DLBCL cells relative to the normal B cell (Figure 3(a)). OCI-ly1 and SU-DHL-4 cells showed relatively higher sensitivity to the dysregulation of HCG18, and therefore these two cells were selected for the following cell experiments. The HCG18-knockdown cell was established through the transfection of HCG18 siRNA, where HCG18 was significantly suppressed (Figure 3(b)).
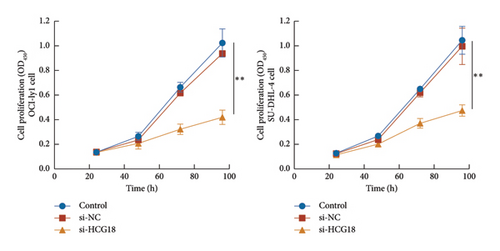
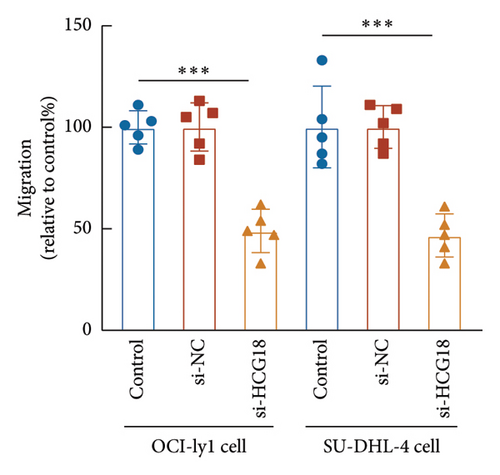
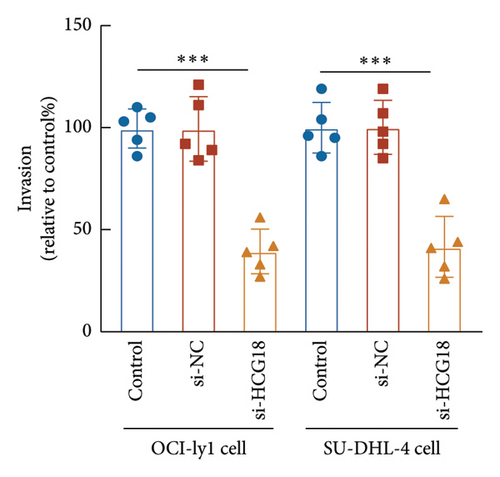
The knockdown of HCG18 significantly suppressed the proliferation of both OCI-ly1 and SU-DHL-4 cells (Figure 3(c)). The dramatical inhibitory effects of HCG18 knockdown were also observed in the migration (Figure 3(d)) and invasion (Figure 3(e)) of OCI-ly1 and SU-DHL-4 cells.
3.3. The Regulatory Mechanism Underlying HCG18 in DLBCL Cells
miR-494-5p was selected from predicted miRNAs from the lncRNA SNP database due to its previously demonstrated function in DBLCL. In OCI-ly1 and SU-DHL-4 cells, the knockdown of HCG18 significantly increased miR-494-3p, which was suppressed by the transfection of miR-494-3p inhibitor (Figure 1(a)). Consistently, the regulation of miR-494-3p could negatively modulate the luciferase activity of HCG18 in OCI-ly1 and SU-DHL-4 cells with several binding sites (Figure 1(b)).
The inhibitory effects of HCG18 knockdown on OCI-ly1 and SU-DHL-4 cell growth (Figure 4(a)) and metastasis (Figures 4(b) and 4(c)) were reversed by the transfection of miR-494-3p inhibitor, indicating the involvement of miR-494-3p in the function of HCG18.
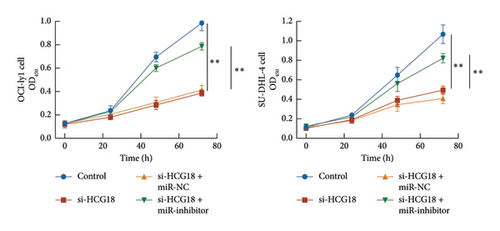
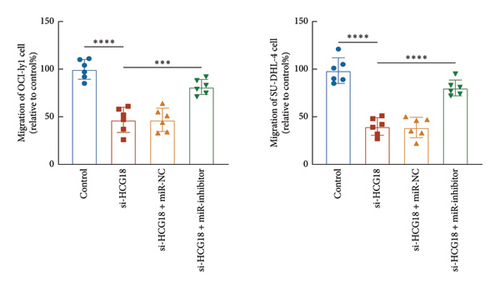
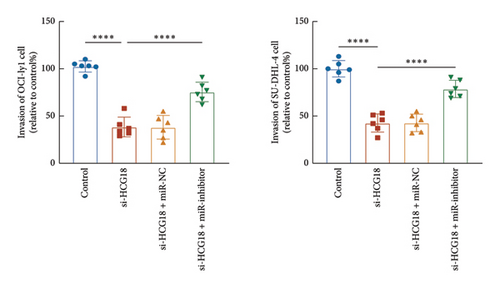
3.4. Prediction of Downstream Regulatory Mechanism of miR-494-3p
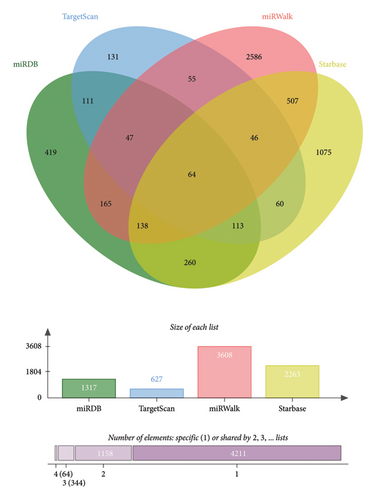
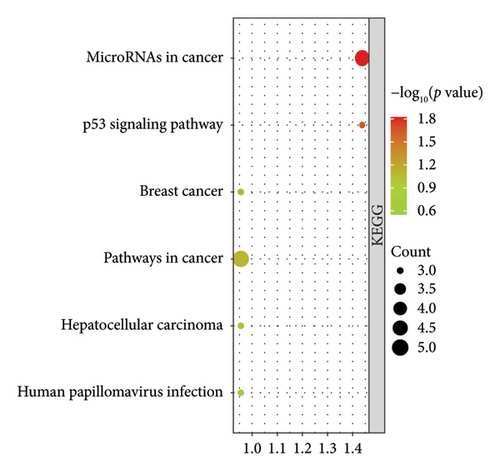
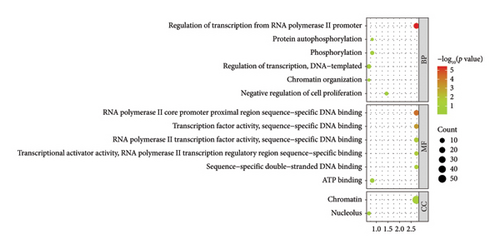
The downstream targets of miR-494-3p were predicted from four public databases, including miRDB (1137 genes predicted), Target Scan (627 genes predicted), miRWalk (3608 genes predicted), and STARBASE (2263 genes predicted), and there were 64 genes enriched in the intersection of the four databases (Figure 5(a)). The function of the enriched genes was annotated from the DAVID database. It was found that enriched genes were mainly correlated with cancer-related pathways, including microRNAs in cancer and pathways in cancer (Figure 5(b)). The biological process (BP), molecular function (MF), and cellular component (CC) classes of Gene Ontology (GO) function annotation mainly involved transcription regulation, DNA binding, and chromatin (Figure 5(c)).
4. Discussion
Due to the high heterogeneity and individual differences, early detection and outcome prediction of DLBCL are still critical challenges in its clinical management [16]. The molecular mechanism underlying the occurrence and development of DLBCL remains unclear, which makes it difficult to provide more targeted and individualized treatment for DLBCL patients. More importantly, the prognosis of DLBCL patients is influenced by various clinical factors, which also increase the difficulty in prognosis prediction. Numerous research has been devoted to improving the diagnosis of DLBCL aiming to improve the clinical prognosis. However, the mortality of DLBCL is still unsatisfactory. Recently, the significance of lncRNAs in cancer onset and progression has attracted numerous attention, and several lncRNAs have been demonstrated to possess great potential in the BP of DLBCL. For instance, lncRNA SNHG14 accelerated the progression and immune evasion of DLBCL via regulating the miR-5590-3p/ZEB1 axis [17]. Downregulated LncRNA SMAD5-AS1 sponged miR-135b-5p and further inhibited the proliferation of DLBCL cells via inactivating Wnt/β-actin signaling [18]. Moreover, there were many studies devoted to establishing lncRNA signatures correlated with the prognosis, recurrence, and other outcomes of DLBCL. Gao et al. formed a 6-lncRNA signature via filtering prognosis-related lncRNAs from GEO and CGA databases. This signature included lncRNA AC244090.1, SNHG26, RPARP-AS1, AC018521.5, AC023590.1, and PRKCQ-AS1 and showed dramatic significance in predicting DLBCL prognosis and risk [19]. A similar bioinformatic study constructed relapse-related lncRNA-mRNA co-expression profiling in Hodgkin lymphoma and identified a series of candidate lncRNA and mRNA biomarkers, where HCG18 was mentioned [14]. Here, a significant elevation of HCG18 expression was observed in tumor tissues of DLBCL patients, which is consistent with its abnormal expression in Hodgkin lymphoma. Upregulation of HCG18 could predict poorer development-free survival of patients and could independently predict the adverse outcomes of DLBCL patients. Ann-Arbor stage and international prognostic index (IPI) scores are commonly used indicators to assess the condition and severity of DLBCL patients [20–23]. The higher expression of HCG18 was associated with the advanced stage and the high IPI score. Therefore, the significance of HCG18 in DLBCL prognosis and development was also evidenced.
Cellular BP are critical factors associated with the tumor progression of malignant cancers. Cell proliferation is related with tumor growth, while cell migration and invasion ability indicated cell metastasis and were associated with the malignancy and development of tumors. Previously, HCG18 was revealed to regulate the cellular processes of osteosarcoma, where the knockdown of HCG18 significantly suppressed the proliferation, migration, and invasion of osteosarcoma [24]. In colorectal cancer, upregulated HCG18 not only facilitated cell growth and metastasis but also enhanced the resistance to cetuximab [25]. The viability, metastasis, and epithelial-mesenchymal transition of papillary thyroid cancer cells were inhibited by HCG18, while cell apoptosis was enhanced, disclosing its tumor suppressor role in papillary thyroid cancer [26]. In the present study, consistent upregulation of HCG18 was observed in DLBCL cells. Through silencing HCG18, its function in cellular processes was disclosed. It was found that the proliferation, migration, and invasion were all suppressed by the knockdown of HCG18 suggesting the promoter role of HCG18 in DLBCL cells.
LncRNAs displayed their function via sponging downstream miRNAs, which are involved in tumor progression. According to the public database, miR-494-3p was predicted to be sponged by HCG18 with several binding sites. The differential expression of miR-494-3p was disclosed in Nasal natural killer cell lymphoma, and DLBCL, miR-494-3p was reported to mediate the promoted effect of lncRNA SBF2-AS1 on cellular processes [15, 27]. Here, miR-494-3p was negatively regulated by HCG18 in DLBCL cells, and miR-494-3p could reverse the effect of HCG18 on DLBCL cell growth and metastasis. Hence, negatively modulating miR-494-3p was hypothesized as the molecular mechanism underlying the function of HCG18 during DLBCL progression. Furthermore, this study performed a prediction of the targets of miR-494-3p, and from the results of function enrichment, it was found that miR-494-3p could target several genes related with cancer pathways, DNA binding, and transcription, which are closely associated with DLBCL progression. Previous studies have also demonstrated the regulation of related pathways by miR-494-3p in human cancers. For example, in endometrial cancer, miR-494-3p was revealed to regulate the PTEN/PI3K/AKT pathway and therefore promote the tumor progression [28]. This regulation was also evidenced to mediate the progression of hepatocellular carcinoma [29]. Additionally, PTEN/PI3K/AKT pathway has been considered to play critical role in the progression of DLBCL and might be associated with rituximab resistance for minority of patients [30, 31]. However, due to financial and time constraints, the deep regulatory mechanism has not been revealed. Whether the PTEN/PI3K/AKT was regulated by miR-494-3p, and whether this kind of regulation was involved in the development of DLBCL have not been revealed. Hence, further mechanism studies are necessary to complete the regulatory network of HCG18 in DLBCL.
However, there are several subtypes of DLBCL according to pathological, immune, and other clinical symptoms [32]. Different subtypes are associated with various risk factors [33]. Therefore, future investigations should refine the subtypes to clarify the role of HCG18 in DLBCL pathogenesis and progression. Due to the relatively low incidence rate of DLBCL, the study enrolled patients meeting the inclusion and exclusion criteria and neglected the calculation of sample size. Moreover, this is a retrospective study with a retrospective analysis of clinical. A prospective study is necessary, and future studies should perform a more rigorous calculation of sample size and expand a larger sample size from multiple-center participants to improve the universality of the clinical results and provide more reliable references. For the involvement and regulatory effect of HCG18 on DLBCL development, there is a lack of in vivo validation, which needs further animal modeling experiments.
5. Conclusion
Taken together, upregulated HCG18 in DLBCL could serve as a biomarker for disease screening and development monitoring. HCG18 also acted as a tumor promoter by targeting miR-494-3p and further regulating functional genes. This study evidenced the involvement of HCG18 in the development of DLBCL and inhibiting HCG18 might be a potential therapeutic strategy for DLBCL management.
Conflicts of Interest
The authors declare no conflicts of interest.
Author Contributions
Huanyu Guo and Yingjie Xie contributed equally to the study.
Funding
No funding was received for this research.
Acknowledgments
The authors have nothing to report.
Open Research
Data Availability Statement
All data generated or analyzed during this study are included in this article. Further enquiries can be directed to the corresponding author.