ERCC3 Gene Associated with Breast Cancer: A Genetic and Bioinformatic Study
Abstract
Female breast cancer is the most common and the fifth deadliest cancer worldwide. It is influenced by a combination of genetic, hormonal, and environmental factors. The excision repair cross-complementation group 3 gene (ERCC3) has recently been identified as a breast cancer susceptibility gene in various cohorts of different geographical and ethnic origin. To explore the role of ERCC3 mutations in breast cancer development and pathological diagnosis, genetic analysis was conducted in 291 patients and 291 controls from mainland China. Bioinformatic analysis and immunohistochemistry (IHC) were performed. A novel ERCC3 mutation p.Y116X was identified in a breast cancer family, while no frequency bias for the genotype and allele of rs754010782 and rs371627165 was observed (all P > 0.05). Bioinformatic analysis revealed that ERCC3 expression was negatively associated with estrogen receptor (ER), progesterone receptor (PR), nontriple-negative status, and nodal status of breast cancers. ERCC3 amplifications and deep deletions primarily occurred in breast invasive cancer not otherwise specified (NOS) and metaplastic breast cancer, respectively. The decreased ERCC3 expression in tumor tissues of patient with p.Y116X mutation was found by IHC. The ERCC3 mutation p.Y116X may increase breast cancer risk in the Han-Chinese population. ERCC3 exhibits potential as a biomarker for the pathological diagnosis of breast cancer.
1. Introduction
Female breast cancer is the most common and the fifth deadliest cancer, accounting for an estimation of 2.26 million newly diagnosed cases and 6.85 million new cancer-related deaths worldwide yearly [1]. It is a heterogeneous disease and its incidence is under the interactive effect of genetic, hormonal, and environmental factors [2]. Breast cancer diagnosed through histopathological analysis in female under the age of 40 years old are defined as early-onset breast cancer which is strongly linked to aggressive triple-negative or human epidermal growth factor 2 (HER2)-positive tumors [3]. Following a four-generation family with multiple female members affected firstly reported in 1866, a series of epidemiological studies recognized genetic as an important risk factor for breast cancer, which explained approximately 5%∼10% of the patients [4]. In recent decades, the advancement of candidate gene-association studies and genome-wide association studies (GWASs), along with meta-analysis of multiple GWAS, has greatly expedited the identification of susceptibility single nucleotide polymorphisms (SNPs), which have been estimated to contribute to approximately 41% of the heritability of breast cancer [5–7]. In addition, more than half of familial cases are of undefined genetic cause [8]. Exploration of the genome of these cases may benefit the genetic underpinnings of breast cancer and bring advances in genetic counselling, cancer-risk assessment, cancer prevention, and patient management.
In recent years, the excision repair cross-complementation group 3 gene (ERCC3) was proposed as a susceptibility gene for breast cancer. Genetic heterogeneity and population-specific frequencies of ERCC3 mutations were observed in cohorts from diverse geographical and ethnic backgrounds [9–12]. To identify ERCC3 mutations that correlate with breast cancer in the Han-Chinese population, genetic analysis of the ERCC3 gene was performed in 291 Han-Chinese breast cancer patients and 291 cancer-free controls by Sanger sequencing, along with a bioinformatic analysis of breast cancer data obtained from various databases.
2. Materials and Methods
2.1. Subjects and Pathological Diagnosis
We enrolled a total of 291 Han-Chinese female patients newly diagnosed with breast cancer (mean age: 48.430 ± 13.822 years). Among them, a cohort of early-onset breast cancers consisted of 97 unrelated sporadic cases and 26 familial patients without BRCA mutations (onset age range: 21∼40 years, at least one close relative diagnosed with breast cancer before the age of 40 or two close relatives diagnosed before the age of 50) [13]. Available relatives of the patients with ERCC3 germline variants were involved for the mutation analysis of pedigree. All patients enrolled in the study were histopathologically confirmed breast cancer based on the 2019 World Health Organization (WHO) classification of tumors of the breast by three independent pathologists from Changsha Hospital for Maternal and Child Health Care, Hunan, China [14]. To create a highly comparable control group, the inclusion criteria were set as Han-Chinese female aged 20∼75 years who have no medical record of cancer. Cases and controls were matched by age (±2 years) at a ratio of 1 : 1. Finally, 291 Han-Chinese female controls with similar age distribution (mean age: 49.113 ± 13.694 years, P = 0.451) were included. This study was approved by the Institutional Review Board of Changsha Hospital for Maternal and Child Health Care, Changsha, Hunan, China (approval number: 2022035).
2.2. DNA Extraction and Quantification
To identify the ERCC3 germline variants in early-onset breast cancers and controls, germline genomic DNA was extracted from peripheral blood leukocytes using QIAamp DNA Blood Mini kit (Qiagen, Germany). For the other breast cancers, ERCC3 germline/somatic variants were detected by extracting genomic DNA from formalin-fixed and paraffin-embedded (FFPE) tumor tissues using the QIAamp FFPE Tissue Kit (Qiagen, Germany). All tumor tissues were fixed in 10% neutral buffered formalin for less than 8 hours. The FFPE blocks preserved within 48 hours were used for DNA extraction. Ten 10-μm FFPE sections of each tumor tissue were manually microdissected, enriching neoplastic cellularity to at least 70%. Following variants identification, germline genomic DNA was also extracted from relatives of early-onset breast cancers with ERCC3 germline variants. The purity of DNA was further evaluated by NanoDrop™ 8000 spectrophotometer (Thermo Scientific, USA). The OD260/280 ratios ranging from 1.8 to 2.0 were deemed acceptable. All procedures were processed referring to manufacturer’s instructions.
2.3. Genetic Analysis
Primers for all exons and exon-intron junctions of the full-length NM_000122.2 mRNA transcript were designed online by using Primer 3 (https://bioinfo.ut.ee/primer3-0.4.0/). All primers were synthesized by Sangon Biotech (Sangon, China) (Table 1). The amplification of the ERCC3 gene through the polymerase chain reaction (PCR) was conducted using the SimpliAmp PCR Thermal Cycler (Thermo Scientific, USA). The PCR conditions consisted of a 3-minute denaturation phase at 98°C, 35 cycles of 10 seconds at 98°C, 10 seconds at 58°C, 15 seconds at 72°C, and a final 5-minute extension phase at 72°C. Each PCR reaction was conducted with a total volume of 25 μL. The reaction mixture consisted of 1 μL of genomic DNA, 1 μL of forward primer, 1 μL of reverse primer, 0.32 μL of deoxyribonucleoside triphosphates mix, 0.15 μL of Taq DNA polymerase, 2 μL of MgCl2, 2.5 μL of 10 × PCR buffer, and 17.03 μL of DNase/RNase-Free distilled water. PCR products were purified using the AxyPrep PCR Clean-up Kit (Axygen, USA) and subsequently subjected to sequencing on an ABI 3500 Genetic Analyzer (Applied Biosystems, USA) employing the BigDye XTerminator Kit.
| Exons | Forward primer (5′ ⟶ 3′) | Reverse primer (5′ ⟶ 3′) | Product size (bp) |
|---|---|---|---|
| 1 | agcggggtcatcttctctct | gtctcccctaggccgagtt | 258 |
| 2 | gggagagatgctggacctg | agctaagggcatgcttacca | 416 |
| 3 | gtggtgttgggcagcttatt | ctccccacaggaaatctgaa | 465 |
| 4 | acccaggcaagaggaagatt | ctgggccacatctcttgttt | 409 |
| 5 | tggtacagatttggcgaagg | tcaagcaatgggtgaagttg | 405 |
| 6 | ccctgctggttctacccttt | gatgccatggctcacagata | 416 |
| 7 | gctttatcccggttgttgac | tccagacacaacagcctgac | 403 |
| 8 | tgtgtgtcgctcagatgtca | ctcagctccagcctgcttac | 473 |
| 9 | gaccagcctgggcaatataa | gcaggtgagcctaagtcctg | 410 |
| 10 | caaactgctggcattacagg | acagccaccttctgcactct | 420 |
| 11 | tgagctcctttttccctctg | caggagaagttccttcagcag | 289 |
| 12 | aaattctggatcagttgacttttt | agaggagtgacctcctgcaa | 227 |
| 13 | cactccagtgcttgcttttg | ggcaaggcctctaatctcct | 342 |
| 14 | gagggaagtgaccaaagcag | ccatccaggcaggactacat | 343 |
| 15 | gactaacatgggctggttcc | gatgcatcttcctcagcaca | 300 |
The coding regions of ERCC3 in breast cancers were sequenced in order to screen for variants. Variant validation was performed on the controls and patients’ relatives by Sanger sequencing on candidate exomes. DNAMAN (version 9) was employed for conducting multiple sequence alignment, while ChromasPro (version 2.1.3) was utilized for sequencing analysis. Variants in this study were designated in accordance with the guidelines of the Human Genome Variation Sequence systematic nomenclature (HGVS, https://www.hgvs.org/). Allelic frequencies of variants were examined by referencing with population databases including the Exome Aggregation Consortium, the 1000 Genomes Project, the Genome Aggregation Database, and the China Metabolic Analytics Project (ChinaMAP).
2.4. Statistical Analysis and Bioinformatic Analysis
The genotypic frequency distribution among the control group was calculated in order to test for Hardy–Weinberg equilibrium using Pearson’s chi-square tests. Differences in genotypic and allelic frequency of variants between patients and controls were compared using Fisher’s exact tests, and a two-sided P value less than 0.05 was considered as statistically significant. Sequence variant interpretations were acquired through the utilization of in silico predictive algorithms including MutationTaster, Polymorphism Phenotyping v2, protein analysis through evolutionary relationships, and functional analysis through Hidden Markov models. The American College of Medical Genetics and Genomics (ACMGs) standards and guidelines for sequence variants interpretation were used to evaluate the functional significance of variants. The structures of the wild-type and mutant proteins were modeled using SWISS-MODEL (https://swissmodel.expasy.org/interactive) and visualized with PyMol (version 2.6.0a0). Relationship between the ERCC3 gene expression and cancer clinicopathological features was analyzed using the Breast Cancer Gene-Expression Miner v4.9 (bc-GenExMiner v4.9, https://bcgenex.ico.unicancer.fr/BC-GEM/GEM-Accueil.php?js=1) online tool. The cBio Cancer Genomics Portal (c-BioPortal, https://cbioportal.org) software was employed to explore genomic alterations of the ERCC3 gene in breast cancer populations.
2.5. Immunohistochemistry (IHC)
The 4-μm sections from FFPE tissues of patients with ERCC3 variants and cancer-adjacent normal breast tissues without ERCC3 alteration were cut, mounted, and deparaffinized. Heat-induced epitope retrieval was performed on sections using sodium citrate buffer (10 mmol/L, pH 6.0) at 110°C for 20 minutes, followed by a 20-minute cooling-off period. The sections were treated with 3% BSA for 10 min to reduce nonspecific staining. Primary antibodies against ERCC3 (XPB rabbit polyclonal antibody from Invitrogen, ThermoFisher, USA) was diluted at a ratio of 1 : 100 and incubated overnight at 4°C. For secondary antibody incubation, MaxVision™ HRP-Polymer anti-Rabbit IHC Kit (MXB biotechnologies, China) was used. The sections were incubated with the secondary antibody at room temperature for 15 minutes. To visualize the antibody-antigen interaction, diaminobenzidine chromogenic procedures were performed using the DAB PLUS Kit (MXB biotechnologies, China) for 4 minutes. Hematoxylin staining was then conducted for 10 seconds. Breast cancer tissues without ERCC3 alteration were selected to serve as positive external control, and negative controls were obtained by replacing primary antibodies with phosphate-buffered saline.
3. Results
Three variants including a novel nonsense variant (c.348C > G, p.Y116X) and two rare missense variants (c.1078C > T, p.R360C, rs754010782; c.1411G > A, p.V471I, rs371627165) were detected in the breast cancers. The frequencies of these variants in the control group were found to adhere to Hardy–Weinberg equilibrium, with all P values exceeding 0.05. The allelic frequencies of the variants in public databases, along with their functional predictions, suggest a possible clinical significance (Table 2). The nonsense variant, c.348C > G (p.Y116X), was identified in the proband (P-1) and not contained in population databases. In her family, the mother was diagnosed with nonspecific invasive carcinoma (grade II) in the right breast at the age of 38. This variant cosegregated with breast cancer in the family by germline mutation analysis (Figures 1(a), 1(b), 1(c), and 1(d)). The pathological findings of breast cancer in the proband and her affected mother were displayed (Figure 2(a) and Supplementary Figure 1). The rs754010782 and rs371627165 were identified in sporadic patients (rs754010782 in S-1 and S2; rs371627165 in S-3, S-4, and S-5).
| Variant | Cases | Samples | Zygosity | dbSNP ID | Allele frequencies | In silico predictions | ACMG | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide change | Amino acid change | ExAC_all | 1000 genomes_all | GnomAD_all | ChinaMAP | MutationTaster | PolyPhen-2 | Panther | FATHMM | |||||
| c.348C > G | p.Y116X | P-1 | PBL | Het | — | — | — | — | — | Disease causing | — | Probably damaging | — | LP |
| c.1078C > T | p.R360C | S-1 and 2 | FFPE | Het | rs754010782 | 3.300 × 10−5 | — | 8.543 × 10−5 | — | Disease causing | Probably damaging | Probably damaging | Tolerated | US |
| c.1411G > A | p.V471I | S-3, 4 and 5 | FFPE | Het | rs371627165 | 2.479 × 10−4 | 2.596 × 10−3 | 2.891 × 10−4 | 7.083 × 10−4 | Disease causing | Benign | Probably damaging | Tolerated | US |
- PBL, peripheral blood leukocytes; FFPE, formalin fixed and paraffin embedded; dbSNP, the single nucleotide polymorphism database; ExAC_all, exome aggregation consortium; gnomAD_all, genome aggregation database; ChinaMAP, the China metabolic analytics project; PolyPhen-2, polymorphism phenotyping version 2; PANTHER, protein analysis through evolutionary relationships; FATHMM, functional analysis through hidden Markov models; Het, heterozygous; ACMG, the American College of Medical Genetics and Genomics; LP, likely pathogenic; US, uncertain significance.
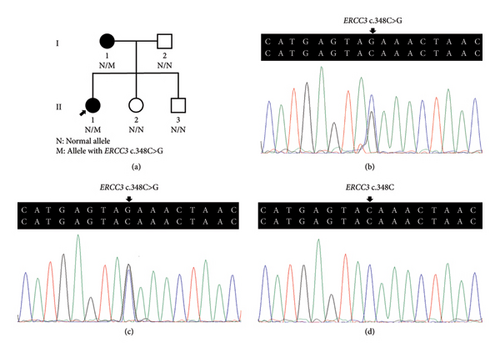
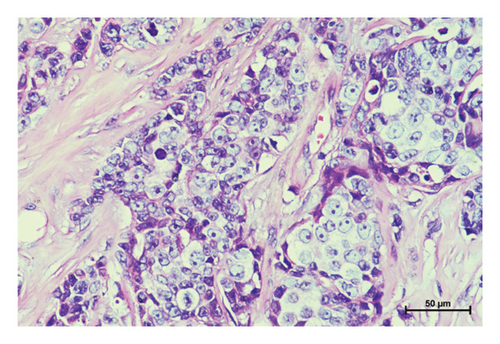
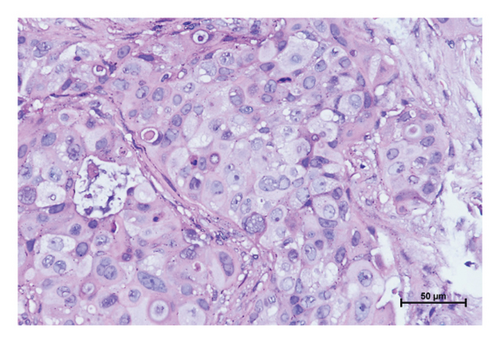

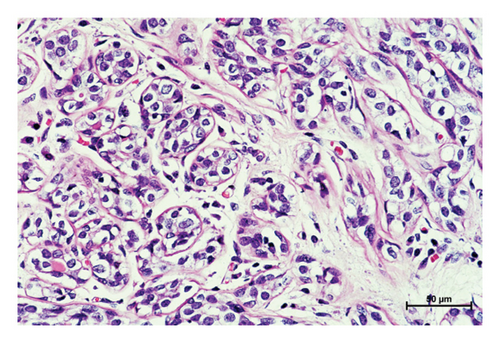
The clinicopathological characteristics of 6 patients were summarized (Table 3 and Supplementary Figures 2 and 3). The distribution of genotypic frequencies and allelic frequencies are presented in Table 4. No statistically significant differences for genotypic and allelic frequencies of rs754010782 (genotype: P = 0.499; allele: P = 0.500) and rs371627165 (genotype: OR = 3.021, 95% CI = 0.312∼29.212, and P = 0.624; allele: OR = 1.503, 95% CI = 0.250∼9.026, and P = 1.000) were observed between the patients and controls. According to the ACMG standards and guidelines, “likely pathogenic” was tagged to p.Y116X while p.R360C and p.V471I were classified as “uncertain significance.”
| Variant | Cases | Onset age | Familial history | TNM stage | Tumor location | Grade | IHC | ||
|---|---|---|---|---|---|---|---|---|---|
| ER | PR | HER2 | |||||||
| p.Y116X | P-1 | 31 | Y | T2N1M0 | Unilateral | III | + | + | − |
| p.R360C | S-1 | 35 | N | T1N0M0 | Unilateral | II | + | − | − |
| S-2 | 38 | N | T1N2M0 | Unilateral | II | + | − | − | |
| p.V471I | S-3 | 62 | N | T3N1M0 | Unilateral | III | + | + | − |
| S-4 | 57 | N | T1N0M0 | Bilateral | I | + | − | + | |
| S-5 | 40 | N | T1N0M0 | Unilateral | II | + | − | − | |
- IHC, immunohistochemistry; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor 2.
| dbSNP ID | Genotype/allele | Patients | Controls | P | OR (95% CI) |
|---|---|---|---|---|---|
| — | CC | 290 | 291 | — | — |
| CG | 1 | 0 | |||
| GG | 0 | 0 | |||
| C | 581 | 582 | — | — | |
| G | 1 | 0 | |||
| rs754010782 | CC | 289 | 291 | 0.499 | — |
| CT | 2 | 0 | |||
| TT | 0 | 0 | |||
| C | 580 | 582 | 0.500 | — | |
| T | 2 | 0 | |||
| rs371627165 | GG | 288 | 290 | 0.624 | 3.021 (0.312∼29.212) |
| GA | 3 | 1 | |||
| AA | 0 | 0 | |||
| G | 579 | 580 | 1.000 | 1.503 (0.250∼9.026) | |
| A | 3 | 2 | |||
- OR, odds ratio; CI, confidence interval.
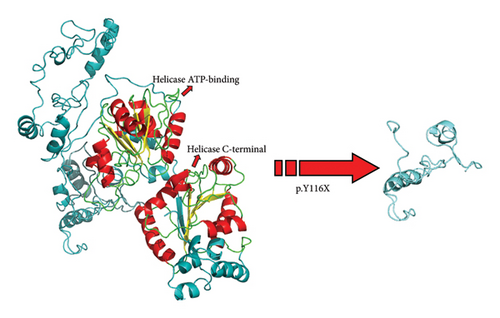
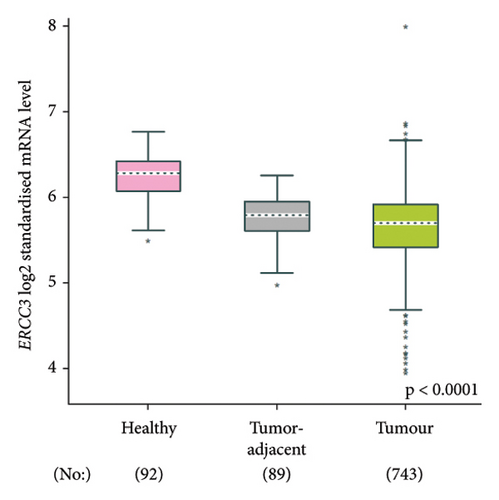
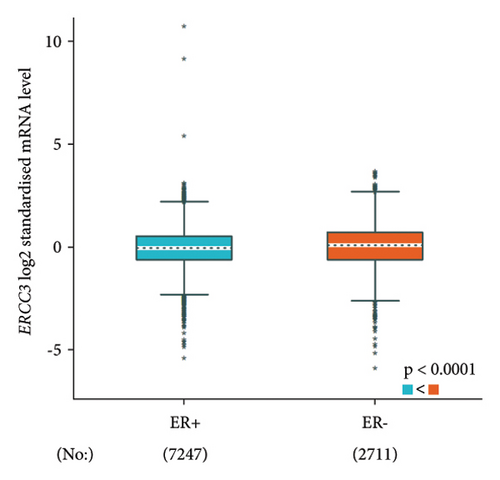
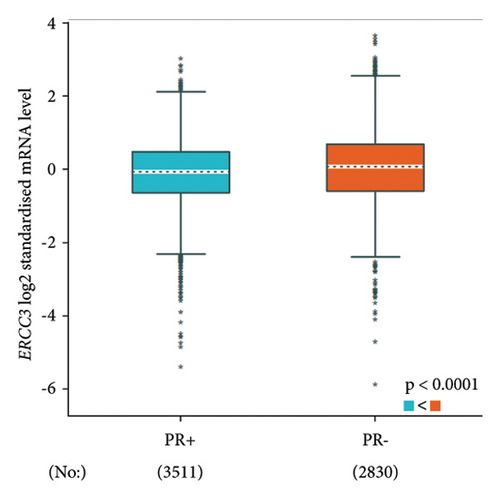
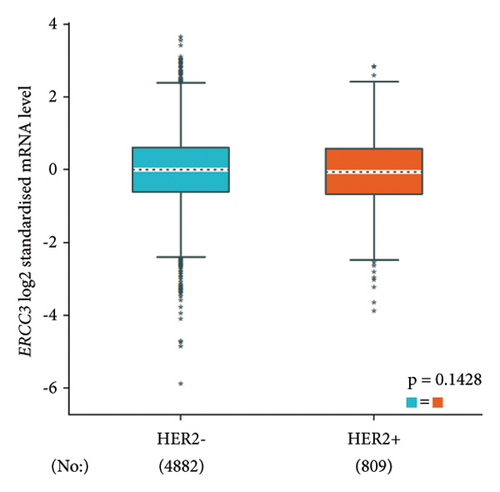
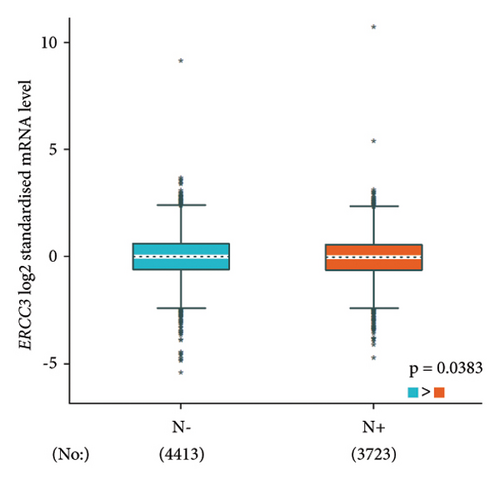
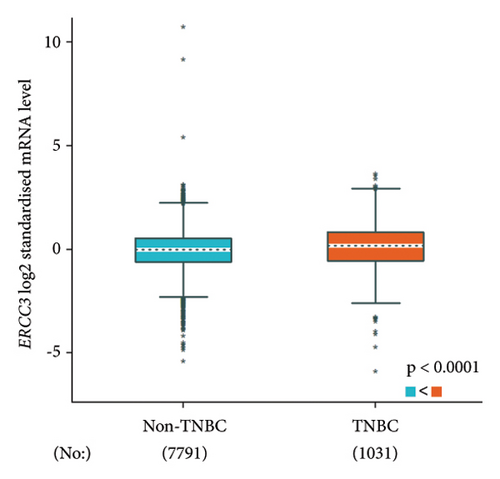
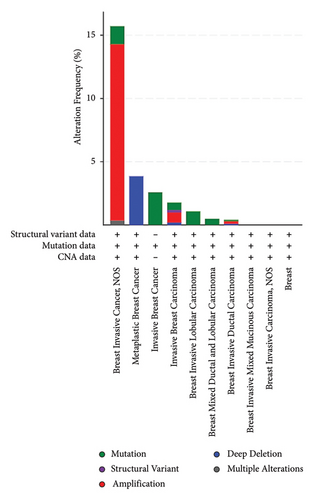
The p.Y116X truncated the ERCC3 protein and caused the loss of helicase ATP-binding domain and helicase C-terminal domain (Figure 3). Analysis of ribonucleic acid sequencing data from the Cancer Genome Atlas and the Genotype-Tissue Expression by bc-GenExMiner v4.9 demonstrated a decrease in ERCC3 expression in tumor tissues compared to both tumor-adjacent and healthy tissues (Figure 4(a)). DNA microarrays data analysis indicated that the estrogen receptor (ER) and progesterone receptor (PR) status had a negative association with ERCC3 expression (Figures 4(b) and 4(c)), and the human epidermal growth factor receptor-2 (HER-2) status seemed to be independent of ERCC3 expression (Figure 4(d)). In addition, decreased ERCC3 expression was observed in patients with positive nodal status (Figure 4(e)). The nontriple-negative breast cancer group also displayed a lower level of ERCC3 expression than the triple-negative breast cancer group (Figure 4(f)). A total of 7101 breast cancer patients from 8 studies were involved in analysis by c-BioPortal, and 1.2% had ERCC3 gene genomic alterations (Figure 5(a)). Amplification of ERCC3 primarily occurred in breast invasive cancer not otherwise specified (NOS), while most of deep deletions occurred in metaplastic breast cancer (Figure 5(b)).

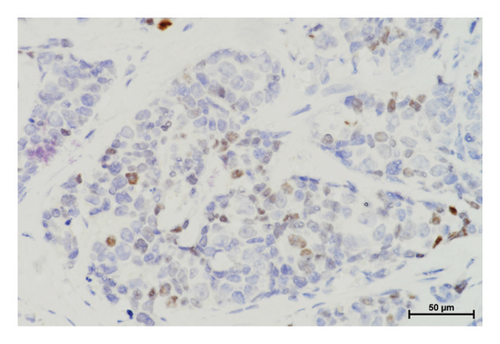
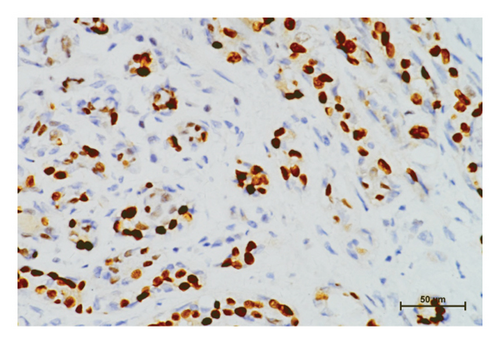
In the tumor tissues of patient P-1, IHC exhibited a faint nuclear staining in <10% of cells (Figure 2(b)). The normal staining of positive external control confirmed that the IHC results were the standard (Figures 2(c) and 2(d)). The IHC staining of normal breast tissues revealed a moderate to strong staining of wild-type ERCC3 expression (Figures 2(e) and 2(f)). Diffuse and mild ERCC3 staining was observed in the tumor tissues of patient S-1, S-2, S-3, and S-5. Diffuse and moderate staining was present in the tumor tissues of patient S-4. IHC analysis indicated an obviously ERCC3 negative in the tumor tissues of the patient with p.Y116X mutation, while in tissues of patients with variants of uncertain significance, mild or moderate staining nucleus in >80% of cells was found (Supplementary Figures 2 and 3).
4. Discussion
The breast cancer susceptibility gene variants are considered the most significant familial risk factor for the occurrence of this disease, particularly in early-onset cases [15]. With the knowledge of today, about a proportion of 5%∼10% breast cancers occur due to genetic risk, and pathogenic variants in breast cancer susceptibility genes are responsible for 25%∼30% hereditary cases [16]. However, over half of hereditary cases without variants in these genes indicate a missing heritability in breast cancer genetics [8]. The ERCC3 gene encodes an ATP-dependent 3′ to 5′-directed DNA helicase that plays a crucial role in basal RNA transcription and the nucleotide excision repair (NER) pathway as the p89 subunit of the human transcription factor IIH (TFIIH) [17, 18]. Patients with ERCC3 mutations manifest NER-defective syndromes including xeroderma pigmentosum group B (OMIM 610651) and trichothiodystrophy 2 (OMIM 616390) and have a high increased risk for skin cancer [19, 20].
Moreover, it has been reported that the ERCC3 gene variants are associated with an elevated risk of developing various types of cancer, including breast cancer, ovarian cancer, lung cancer, osteosarcoma, bladder cancer, colorectal cancer, medulloblastoma, chronic lymphocytic leukemia, malignant pleural mesothelioma, and thyroid cancer (Table 5) [9–12, 21–32, 34]. Multiple multilocus inherited neoplasia alleles syndrome cases with ERCC3 truncating mutations were reported, which implied that ERCC3 variants also possibly play a role in modifying the cancer phenotype [11, 31, 35]. In vitro study suggested that the recurrent mutation, c.325C > T (p.R109X), can cause reduced ERCC3 expression and impaired DNA-repair function [9].
| Nucleotide change (NM_000122.2) | Amino acid change (NP_000113.1) | Chromosome location (hg38) | dbSNP ID | Related-cancer |
|---|---|---|---|---|
| c.325C > T [9–11] | p.R109X | chr2: 127292756 | rs34295337 | Breast cancer |
| c.335dup [11] | p.H112QfsX4 | chr2: 127292746 | — | Breast cancer, ovarian cancer |
| c.471 + 555A > G [21, 22] | — | chr2: 127292055 | rs4150407 | Bladder cancer, lung cancer |
| c.583C > T [11] | p.R195X | chr2: 127289763 | — | Breast cancer, ovarian cancer |
| c.658-1G > A [11] | — | chr2: 127289502 | — | Breast cancer |
| c.694A > G [23] | p.T232A | chr2: 127289465 | — | Lung cancer |
| c.700C > T [12, 24] | p.R234X | chr2: 127289396 | — | Breast cancer |
| c.760C > T [11] | p.Q254X | chr2: 127289399 | — | Ovarian cancer |
| c.786_791delAGAAGA [25] | p.E262_E264del | chr2: 27289368_127289373 | — | Malignant pleural mesothelioma |
| c.917T > C [26] | p.I306T | chr2: 127288770 | — | Colorectal cancer |
| c.1300G > T [27] | p.E434X | chr2: 127286745 | — | Medulloblastoma |
| c.1342 + 2179G > A [28] | — | chr2: 127284524 | rs4150434 | Lung caner |
| c.1343−2708A > G [29] | — | chr2: 127283339 | rs4150441 | Osteosarcoma |
| c.1346A > G [23] | p.K449R | chr2: 127280628 | — | Lung cancer |
| c.1354C > T [12] | p.R452X | chr2: 127280620 | — | Breast cancer |
| c.1421_1422insA [11] | p.D474EfsX2 | chr2: 127280553 | rs587778281 | Breast cancer |
| c.1720C > T [12] | p.R574X | chr2: 127279183 | — | Breast cancer |
| c.1757delA [10, 30, 31] | p.Q586RfsX25 | chr2: 127272935 | — | Breast cancer |
| c.1757_1758delAG [11, 32] | p.Q586RfsX17 | chr2: 127272934_127272935 | — | Breast cancer, ovarian cancer, thyroid cancer |
| c.1841C > A [12] | p.S614X | chr2: 127271440 | — | Breast cancer |
| c.1854_1867del [33] | p.E619HfsX24 | Chr2: 127271416_127271429 | — | Breast cancer |
| c.2111C > T [34] | p.S704L | chr2: 127259402 | rs4150521 | Chronic lymphocytic leukemia |
| c.2218-1G > A [11] | — | chr2: 127257728 | — | Breast cancer |
In this case-control study among a Han Chinese population composed of 592 participators, we assessed the role of ERCC3 variants including a novel germline nonsense variant c.348C > G (p.Y116X) and two missense variants rs754010782 (p.R360C) and rs371627165 (p.V471I) in breast cancer pathogenesis. The c.348C > G (p.Y116X) cosegregated with breast cancer in the family. Previous exome sequencing analysis of the proband excluded the BRCA mutations, suggesting that this mutation is the most likely genetic cause for this breast cancer family. No frequency bias for the genotype and allele of rs754010782 and rs37162716 was observed in our well-characterized cohort. The truncating mutation p.Y166X is found to potentially increase the risk of breast cancer in mainland Han-Chinese population, while the other two missense variants, rs754010782 and rs371627165, have no apparent association with the development of breast cancer. Some variables, including constraints on the sample size, no matching germline DNA, FFPE-DNA damage, and the infrequency of the genotypes or alleles among the Han-Chinese population, may contribute to the inconclusive outcomes of the two variants rs754010782 and rs371627165. In addition, intrinsic and extrinsic factors, such as epigenetics, geographical origin, and limited sample size, may lower the representativeness of this Han-Chinese population. Further replication studies in a large number of familial breast cancers may provide more genetic evidence for the risk role of p.Y166X mutation.
Up to now, only 13 ERCC3 truncating and splicing mutations have been reported in a minimum of 29 breast cancer patients and confer a 1.53 ∼ 3.54-fold breast cancer risk [9–12, 24, 30, 31, 33, 36, 37]. Impaired ERCC3 function in NER may be a potential pathogenesis for breast cancer [9]. In the analysis conducted using bc-GenExMiner v4.9, it was observed that a decrease in ERCC3 expression is significantly associated with ER, PR, and nontriple-negative status, as well as the nodal status. Studies have also reported a significantly higher frequency of ERCC3 mutation in ER-positive breast cancer [9, 12]. Confounding factors, including the geographical and ethnic backgrounds of the study population, methodological variations, potential interactions with other gene variants, and environmental factors, as well as additional complicating factors, are overlooked during the analysis. Therefore, they should all be taken into account when interpreting the results of this bioinformatic analysis.
By IHC analysis, ERCC3 negative expression was identified in the tumor tissues of patients with p.Y116X mutation. The c.348C > G (p.Y116X) mutation may be involved in breast cancer by causing reduction in ERCC3 expression. This result is consistent with previous studies that ERCC3 truncating mutation cause decreased ERCC3 expression [9]. Mild or moderate expression was observed in tumor tissues of patients with rs754010782 and rs371627165 variants. The tumor tissues of patients S-1, S-2, S-3, and S-5 tend to present a slightly decreased ERCC3 expression. The patient S-4 has the similar ERCC3 expression level compared with positive external control and normal breast tissues. Influences of rs754010782 and rs371627165 on ERCC3 expression requires further evidence from more variant carriers.
ERCC3 truncating mutation has the potential to serve as a biomarker for the pathological diagnosis of breast cancers. By exploring ERCC3 gene genomic alterations in breast cancers using c-BioPortal, we found that ERCC3 amplification concentratedly occurred in breast invasive cancer NOS, and most of deep deletions occurred in metaplastic breast cancer. ERCC3 copy number variant (CNV) detection may help confirm histopathological type of breast cancer and breast cancer metastasis.
5. Conclusion
In conclusion, this study revealed that ERCC3-truncating mutation p.Y166X may contribute to the breast cancer development in Han-Chinese population. Since the absence of common variant with a high allele frequency in our study, more genetic analysis studies in larger cohorts from different regions as well as functional studies are warranted to estimate the relative risk for breast cancer conferred by ERCC3 mutations. Our findings also imply that ERCC3-truncating mutation and CNV have potential clinical significance in pathological diagnosis of breast cancer.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81903199), the Natural Science Foundation of Hunan Province (2021JJ40373), and Hospital-Level Research Project of Changsha Hospital for Maternal and Child Health Care (2022YJ02).
Open Research
Data Availability
The data used to support the findings of this study are available from the corresponding author upon reasonable request.