Intellectual Disability and Blended Phenotypes: Insights from a Centre in North India
Abstract
Intellectual disability (ID) is seen in around 2.5% of global population and can vary from mild to severe and profound ID. There can be multiple affected family members if it is inherited, though many autosomal dominant ID cases would be due to de novo mutations are very less likely to recur in families. A confirmatory diagnosis is facilitated by genetic testing like chromosomal microarray and next generation sequencing. We describe here our cohort of 15 patients: children and adolescents with ID diagnosed by using sequencing technologies and parental segregation studies. Most of the variants identified were de novo variants and consistent with sporadic occurrence, and blended phenotypes were identified. Appropriate genetic counseling was performed and options for prenatal diagnosis were discussed. Thus, advanced sequencing technologies enable identification of likely causative de novo variants associated with intellectual disability and dysmorphism.
1. Introduction
Intellectual disability is a clinically and genetically heterogenous group with multiple tests being used to reach a definitive diagnosis in complicated cases [1–3]. The presentations include developmental delay, autism, seizures, behavioral problems, and magnetic resonance imaging (MRI) abnormalities in certain cases. With availability of high-throughput technologies, more cases are getting molecular confirmation and better prognostication. The traditional Sanger sequencing using fluorescently labelled terminating nucleotides followed by gel electrophoresis has been the gold standard for sequencing. The major advantage of new “second generation sequencing” (SGS) technology is considerably high throughput and hence low cost per base sequenced. In the SGS technology developed by Illumina, adapter-ligated DNA sequences are amplified directly in the flow cell subsequently utilised for sequencing. In SOLiD (sequencing with oligonucleotide ligation and detection) technologies, adapter-ligated fragments are hybridized to beads on glass slide, coated with an oligonucleotide complementary to one of the adapters and used for amplification in a water-in-oil emulsion polymerase chain reaction (PCR). Thus, next generation sequencing (NGS) is being increasingly used for confirmation of diagnosis in phenotypes with genetic heterogeneity. We report some Indian patients with intellectual disability identified on NGS based testing.
2. Methods
This is an analysis of the patients seen in the specialty clinic of tertiary care centre. The main objective was to find variants especially de novo single nucleotide variants (SNVs) related to ID and phenotypic variability in Asian Indians. The clinical evaluation was done as per standard protocols including X-rays of the dorso-lumbar spine and wrist for identifying vertebral abnormalities, dysostosis, and advanced or delayed bone age. Ultrasonography of abdomen was done to identify any renal anomalies, and echocardiography was done to find out cardiac anomalies. In unexplained ID, basic metabolic work up like plasma ammonia, serum lactate and creatine kinase levels were performed. The inclusion criteria included (1) presence of developmental delay, microcephaly, and/or behavioural abnormalities; (2) negative results on karyotyping and multiplex ligation probe amplification (MLPA) and/or chromosomal microarray testing not showing copy number variants; (3) referred to clinic for dysmorphism or other anomalies; and (4) MRI not suggestive of structural anomalies or neurometabolic disorder.
NGS for targeted genes causing intellectual disability or autism was done as per standard protocols using variant filtering, prioritization, etc. Variant classification was done as per American College of Medical Genetics and Genomics (ACMG) guidelines [4]. Pretest and posttest genetic counseling was offered in the families. Ethical clearance was obtained from the Institute ethics committee. Written informed consent was taken from the parents for genetic testing, and for photographs from all the participants, and Indian Council of Medical Research (ICMR) ethical guidelines were followed.
3. Results
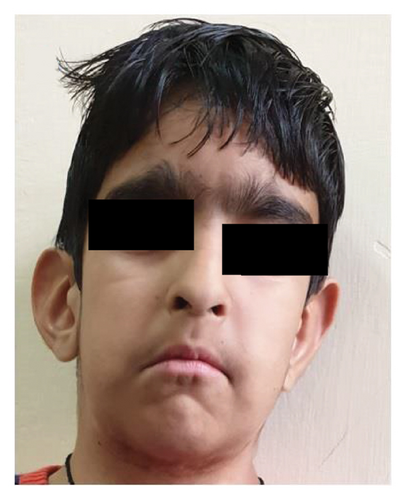
This study encompasses fifteen patients with ID, in a majority of whom heterozygous variants were identified using targeted NGS. MRI brain was done in Case 2 (Figure 1) at 1 year age and showed nonspecific altered signal intensities and a small arachnoid cyst. Figure 1 depicts facial profile of patients with Coffin Siris syndrome–patients 2 and 13 in Table 1. Table 1 lists the clinical details and variants found in these patients. Figure 1S shows Sanger depicting variant in Case 2. Four of these cases are described in detail below:

| Sl. no. | Age/Sex | Clinical features | Diagnosis | Gene | Zygosity | HGVSc | HGVSp | Variant classification |
|---|---|---|---|---|---|---|---|---|
| 1 | 4 year/F | Developmental delay, seizures, microcephaly, hypotonia, epicanthic folds, brachydactyly | Rett syndrome | MECP2 | Heterozygous | NM_001110792.2: c.352C > T | NP_001104262.1: p.Arg118Trp | Pathogenic |
| 2 | 5 year/F | Microcephaly, developmental/speech delay, hypotonia, hirsutism, preauricular sinuses, short little fingers, pubic hairs | Coffin Siris syndrome | ARID1B | Heterozygous | NM_001374820: c.5467delG | NP_065783.3: p.Gly1824ValfsTer27 | Likely pathogenic |
| 3 | 4 year/F | ID, obesity, abnormal behaviour | Dominant ID | PDE4D | Heterozygous | NM_001104631.2: c.701G > A | NP_001098101.1: p.Arg234Gln | VUS |
| 4 | 6 year/M | Developmental delay, microcephaly, behavioural problems, bilateral peritrigonal hyperintensities | Dominant ID | MYT1L | Heterozygous | NM_015025.4: c.1700G > A | NP_055840.2: p.Arg567Gln | Likely pathogenic |
| 5 | 13 year/M | ID, hyperactivity, large ears, bulbous nose, broad eyebrows, hemivertebrae (L4-L5) | KBG syndrome | ANKRD11 | Heterozygous | NM_001256182.2: c.7814T > G | NP_001243111.1: p.Leu2605Arg | Likely pathogenic |
| 6 | 5 year/F | Coarse facies, autism, intellectual disability, no malformations | Sporadic ID | AUTS2 | Heterozygous | NM_015570.4: c.416_419delAGAG | NP_056385.1: p.Lys146LeufsTer61 | Likely pathogenic |
| 7 | 3.5 year/M | Developmental delay, ID, squint | X-linked ID, Billuart type | OPHN1 | Hemizygous | NM_002547.3: c.2253_2262delAGCTACTCCC | NP_002538.1: p.Ala752fs | Likely pathogenic |
| 8 | 1 year/F | Plagiocephaly, squint, delayed milestones, hypotonia | PURA syndrome | PURA | Heterozygous | NM_005859.5: c.791G > T | NP_005850.1: p.Trp264Ter | Likely pathogenic |
| 9 | 3 year/M | Developmental delay, speech delay, microcephaly, happy demeanour, long eyelashes | Schuurs–Hoeijmakers syndrome | PACS1 | Heterozygous | NM_018026.4: c.607C > T | p.Arg203Trp | Pathogenic |
| 10 | 2 year/F | Developmental delay, hyperactivity, triangular facies, microcephaly, bushy eyebrows, anteverted nares, hirsutism, clinodactyly, flat feet | ID_Dual diagnosis | ANKRD11 | Heterozygous | NM_001256183.2: c.1388_1389del | p. Lys463ArgfsTer29 | Likely pathogenic |
| EP300 | Heterozygous | NM_001429.4: c.4261dup | p.Tyr1421LeufsTer22 | Pathogenic | ||||
| 11 | 10 year/M | Intellectual disability, ADHD, dysmorphism, microcephaly, thick lips, ring dermoid | ID with dysmorphism | PITX2 | Heterozygous | NM_001204398.1: c.185–1477C > G | — | VUS |
| 12 | 4 year/F | Developmental delay, autism, toe walking, stereotypic movements of hands | Atypical Rett syndrome | MECP2 | Heterozygous | NM_001110792.2: c.433C > T | p.Arg145Cys | Pathogenic |
| 13 | 8 year/M | Mild intellectual disability, seizures, large ears, coarse facies, synophrys, broad thumbs, club foot | Coffin Siris syndrome | ARID1B | Heterozygous | NM_ 020732.3: c.172G > A | p.Ala58Thr | VUS |
| 14 | 1 year/F | Developmental delay, hypotonia, synophrys, cupped ears, sparse hair, protruding tongue, clinodactyly, hirsutism | Wiedemann-Steiner syndrome | KMT2A | Heterozygous | ENST00000534358.8 chr11: g.118473930C > T | p.Ser924Leu | Likely pathogenic |
| AUTS2 | Heterozygous | ENST00000342771.10 | p.His535_Thr542del | VUS | ||||
| 15 | 3 year/F | Seizures, autism, cardiac defect-VSD, growth retardation, microcephaly, dysmorphism | Angelman like syndrome | GABRA2 | Heterozygous | NM_000807.4: c.455G > A | p.Gly 152Glu | VUS |
| FLNA | Heterozygous | NM_001110556.2: c.6737C > T | p.Pro2246Leu | Likely pathogenic |
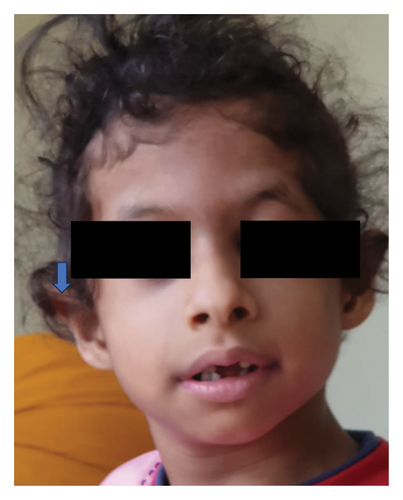
1-year-old female child presented with developmental delay and seizures. Examination revealed plagiocephaly, squint, prominent cheeks, poorly formed nasal columella, inverted V-shaped upper lip, open mouth (Figure 1), and hypotonia. She also had stranger anxiety. The creatine phosphokinase level was 348.5 units (normal level < 228 units), and MRI brain was normal. The thyroid stimulating hormone (TSH) level was 2.58 μIU/ml (normal: 0.7–5.97 units). NGS was done to cover genes causing hypotonia also including congenital myopathy genes. It revealed an exon1 variant in the PURA gene. The heterozygous nonsense variant identified was PURA: c.791G > A: p. (Trp264∗). In addition, there were compound heterozygous missense variants in the HERC2 gene: p.Asn370Ser and p.Val2532Leu in exon 10 and exon 47 of the gene.
A two-year-old girl was brought to the outpatient clinic with global developmental delay and hyperactivity. There was no positive family history and no consanguinity. Examination revealed short stature, triangular facies, microcephaly, coarse facies, bushy eyebrows, anteverted nares, clinodactyly, and generalised hirsutism. Some of the features like microcephaly and short stature were consistent with Rubinstein-Taybi syndrome (RSTS) whereas coarse facies, thick eyebrows, and hirsutism were suggestive of possible chromatinopathy like KBG syndrome. The targeted NGS testing revealed two pathogenic variants associated with ID in ANKRD11 and EP300 genes (Table 1), confirming a blended phenotype. Figure 2 depicts facial profile of patient with Coffin Siris syndrome—patient 2 in Table 1.
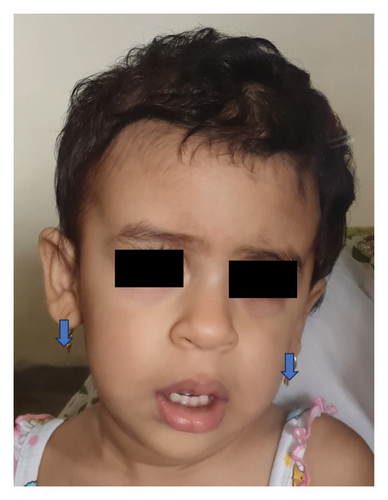
A three-year-old female child was brought with features of seizures, growth deficiency, cardiac defect, and autism. Examination revealed microcephaly and dysmorphism (Figure 3). There were no renal anomalies on ultrasonography. Testing for chromosomal deletions and duplications was negative. Targeted next generation sequencing revealed GABRA2 and FLNA variants. The facies was consistent with that associated with FLNA gene variant, but further characterisation needed.
A 3-year-old male child (case 9) presented with low birth weight (2 kg), developmental delay, microcephaly (−3.4Z), right microcornea, long eyelashes, squint, retrognathia, low-set ears hearing impairment, and a happy demeanor (Figure 2S). Echocardiography revealed 4 mm patent ductus arteriosus (PDA), with left to right shunt. MLPA testing for chromosomal deletions and duplications was negative. NGS testing revealed a likely pathogenic heterozygous variant in PACS1: c.607C > T(p.Arg203Trp).
4. Discussion
Children with ID can have variable presentations, and some have additional phenotypic abnormalities which aid in the diagnosis. Having a child with severe ID in family can create management issues in the family and can be a burden on the public health system especially if additional systemic abnormalities are also present like cardiac or renal anomalies. The usual first line test in unexplained developmental delay or ID with or without dysmorphism is chromosomal microarray (CMA) for deletions and duplications [5]. However, those negative on CMA or having characteristic features of a particular syndrome like Coffin Siris syndrome can be further tested by NGS. MLPA for common deletions or duplications is another cheaper alternative test used for known chromosomal microdeletions or duplications. Nowadays, targeted NGS is being increasingly used for diagnosis of genetic disease, unexplained phenotypes, and for gene expression profiling and genome research.
Intellectual disability can be very heterogenous with overlapping phenotypes. “Overlapping phenotypes” refer to presentation with developmental delay, behavioural abnormalities, with or without seizures or stereotypies in several neurodevelopmental disorders. Few have additional malformations, and MRI brain can show nonspecific abnormalities. In selected cases, a facial gestalt can help in making a clinical diagnosis. Sometimes additional features can help in identifying a particular syndrome as in case of patient 4 and patient 10 in present report. We identified some novel variants in known genes causing ID in the Asian Indians. A variant of unknown significance as per ACMG guidelines can be intriguing and further evaluation becomes important. A parental segregation analysis and use of bioinformatic tools for predicting effect of the variants are being increasingly used for finding out the relevance of the variants identified.
In present analysis, most variants were heterozygous mutations mostly de novo; and 9 variants were pathogenic or likely pathogenic variants. Case 1 showed a heterozygous variant MECP2 (NM_001110792.2): c.352C > T (p.Arg118Trp), rs28934907, located at ChrX:153297719 (GRCh37); pathogenic as per ACMG guidelines, causes Rett Syndrome and is extensively published [6]. The syndrome has X-linked dominant (XLD) inheritance including atypical and preserved speech variants [OMIM #312750]. Another girl in present series with ID and stereotypic hand movements also showed MECP2 gene variant, which has also earlier been reported with patients diagnosed with Rett syndrome.
Case 2 showed a heterozygous null variant (frame-shift) NM_001374820: c.5467delG, in gene ARID1B. It is autosomal dominant disorder and associated with Coffin-Siris syndrome type1 [OMIM #135900] [7]. This variant is not found in gnomAD exomes or gnomAD genomes but has been reported at ClinVar and classified as “Pathogenic” in association with Coffin-Siris syndrome [Accession: SCV001736716.1].
Case 3 showed a heterozygous variant NM_001104631.2: c.701G > A, rs766490621, in PDE4D gene. This VUS is located at Chr5:58481072 (GRCh37). It is reported across population databases with the following frequencies: T = 0.000016 (4/248010, GnomAD_exome); T = 0.000033 (4/119640, ExAC); and T = 0.00003 (1/32060, ALFA).
The MYT1L gene variant in Case 4 is a heterozygous SNV: NM_015025.4: c.1700G > A, rs878853045. It is located at Chr2:1915795 (GRCh37). It is reported at ClinVar [Accession: VCV000235469.8] as “likely pathogenic” in association with autosomal dominant ID. It has previously been reported as a de novo variant in a patient with intellectual disability, plagiocephaly, 5th finger clinodactyly, an ataxic gait, hyperphagia, and cerebral atrophy [reported as p.Arg569Gln] [8].
A male child with ID (Case 5) showed a heterozygous variant NM_001256182.2: c.7814T > G, rs1131691512, in ANKRD11 gene. It is located at Chr16: 89335064 (GRCh37). It is reported at ClinVar [Accession: VCV000429654.4]. This variant is not found in gnomAD exomes or gnomAD genomes. Macrodontia involving upper central incisors is a diagnostic hallmark of KBG syndrome, which is named using initials of first described patients with the syndrome. This condition can also result from microdeletion of chromosome 16q24 in some patients, a copy number variant (CNV), which can be identified on chromosomal microarray [9].
Interestingly, in Case 6 with coarse facies, a heterozygous null variant (frame-shift) was identified NM_015570.4: c.416_419delAGAG, in gene AUTS2. This likely pathogenic variant is associated with autism spectrum disorder due to AUTS2 deficiency. Case 7 showed a hemizygous deletion chrX-67273549-GGGAGTAGCT (10 bp) (OPHN1: p.A752Kfs ∗82). This variant NM_002547.3: c.2253_2262delAGCTACTCCC was not found in gnomAD exomes or gnomAD genomes and not reported at ClinVar; thus the variant was labelled as “likely pathogenic.” OPHN1-related ID is a X-linked recessive disorder affecting males [10].
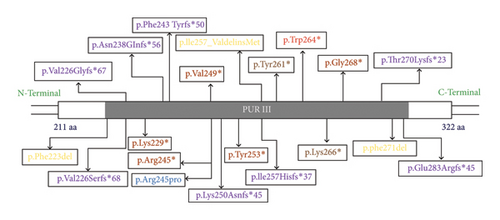
Case 8 showed a heterozygous missense variant, a novel likely pathogenic variant_ NM_005859.5: c.791G > T (p.Trp264∗) in the PURA gene. PURA gene codes for PUR-alpha, which is a purine-rich protein and essential for normal brain development including oligodendrocytes and astrocytes. It is in exon 3 of transcript ENSG00000185129.5 for gene PURA [GTExVersion: v8] in the hotspot region of the gene. In a large cohort study by Reijnders et al., four variants in PURA gene were identified as recurrent variants which include p.Lys97Glu, p. Phe233del, p.Arg245Pro, and p.Phe271del. This disorder has some similarities to cutis laxa and myopathic facies, gait abnormalities and increased creatine kinase suggest muscle involvement in PURA syndrome [11, 12]. The pathogenic and likely pathogenic variants in the PURA gene product, in domain III have been illustrated in Figure 4. A recent article has identified high frequency of de novo mutations or variants (DNMs) in few genes which include PURA, SATB2, SCN1A, and TUBA1A and these are mostly found in cases of neurodevelopmental disorders (NDDs) [13].
Case 9 showed a heterozygous variant, NM_018026.4(PACS1): c.607C > T, rs398123009, is located at 11: 65978677 (GRCh37). It is reported at ClinVar [Accession: VCV000039581.39] where it is classified as “pathogenic:” in association with Schuurs–Hoeijmakers syndrome [14]. The nucleotide position is conserved across 35 mammalian species (GERP RS: 4.79). Blended phenotype was observed in Case 10, a 2-year-old girl with ID. She showed a heterozygous frameshift variant on NGS: NM_001256183.2 (ANKRD11): c.1388_1389del (p.Lys463Argfs ∗29). A “Likely pathogenic” variant causing KBG syndrome. This patient also showed a heterozygous null variant, EP300 (NM_001429.4): c.4261dup (p.Tyr1421Leufs ∗22); associated with Rubinstein-Taybi syndrome (RSTS), a novel pathogenic variant as per ACMG guidelines. Variants in EP300 have also been reported in association with colorectal cancer, and Menke–Hennekam syndrome 2. It is not found in gnomAD exomes or gnomAD genomes. Thus, the NGS testing enabled identification of a “blended” phenotype and hence enabled appropriate genetic counseling. When different variants are causing similar phenotypes, sometimes it may be difficult to differentiate which clinical feature is due to which variant, thus the phenotype is mixed or blended.
Similarly, in case 15, a single variant could not explain the full phenotype. Some features were explained by the FLNA gene reverse phenotyping; but autism and seizures could be explained by the GABRA2 variant; thereby suggesting possibility of a blended phenotype. The FLNA variant c.6737C > T (p.Pro2246Leu), was not found in ExAC and 1000 genomes database, and classified as likely pathogenic using in-silico tools and ACMG guidelines, predicted deleterious on PROVEAN, LRT, and REVEL databases. Franklin automated classification classified it as VUS, leaning towards pathogenic. The GABRA2 variant in this child has earlier been reported as VUS on ClinVar.
The heterozygous variant in Case 11 NM_001204398.1 (PITX2): c.185–1477C > G, rs1578453824, located at Chr4:111544002 (GRCh37). It is neither reported at ClinVar nor in gnomAD exomes or gnomAD genomes. This gene has been earlier associated with ring dermoid which was seen in the child [15]. Thus, the variant was a variant of unknown significance (VUS). Confirmation of SNVs and parental segregation is done by Sanger sequencing. Nowadays, copy number variants (CNVs) can also be identified on NGS analysis by using specialised tools or pipelines due to availability of better technologies or tools using artificial intelligence. Severe intellectual disability with microcephaly and multiple malformations is more likely due to chromosomal cause and hence CMA is first line genetic testing [5].
Thus, we found some known and novel variants in children with ID. Hence, to conclude, NGS enables finding mutations in known genes with relative ease, thus facilitating appropriate genetic counseling in the affected families. It is a useful adjunct to MLPA and CMA to make definitive clinical diagnosis, in patients with mild to moderate intellectual disability.
Ethical Approval
The study and genetic evaluation was as per Declaration of Helsinki. IEC clearance was taken from the Institute Ethics Committee.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors’ Contributions
Data collection for analysis was done by IP and DK. Diagnosis, evaluation, and follow up were done by IP and PK. Molecular findings interpretation was done SVK and SR. Drafting of manuscript was done by IP and assisted by SVK. All authors have read and approve of the manuscript.
Acknowledgments
The authors acknowledged ICMR [2022-NCD II dated 20.09.2023].
Open Research
Data Availability
The data that support the findings of this study are available on request from the corresponding author. One ARID1B variant details are available on ClinVar_ Accession: SCV001736716.1. The data for other patients are not publicly available due to privacy or financial or ethical restrictions.




