A Novel Approach for Best Parameters Selection and Feature Engineering to Analyze and Detect Diabetes: Machine Learning Insights
Abstract
Humans are familiar with “diabetes,” a chronic metabolic disease that causes resistance to insulin in the human body, and about 425 million cases worldwide. Diabetes is a hazard to human health since it can gradually cause significant damage to the heart, blood vessels, eyes, kidneys, and nerves. As a result, it is critical to recognize diabetes early on to minimize its negative consequences. Over the years, artificial intelligence (AI) technology and data mining methods are playing a crucial role in detecting diabetic patients. Considering this opportunity, we present a fine-tuned random forest algorithm with the best parameters (RFWBP) that is used with the RF algorithm and feature engineering to detect diabetes patients at an early stage. We have employed several data processing techniques (e.g., normalization, conversion into numerical data) to raw data during the prepossessing phase. After that, we further applied some data mining techniques, adding related characteristics to the primary dataset. Finally, we train the proposed RFWBP and conventional methods like the AdaBoost algorithm, support vector machine, logistic regression, naive Bayes, multilayer perceptron, and a regular random forest with the dataset. Furthermore, we also utilized 5-fold cross-validation to enhance the performance of the RFWBP classifier. The proposed RFWBP achieved an accuracy of 95.83% and 90.68% with and without 5-fold cross-validation, respectively. Moreover, the proposed RFWBP is compared with conventional machine learning methods to evaluate the performance. The experimental results confirm that the proposed RFWBP outperformed conventional machine learning methods.
1. Introduction
1.1. Background
Diabetes mellitus (DM) is the most common long-lasting noncommunicable public health concern that causes serious health complications, e.g., kidney disease, cardiovascular disease, and lower-limb amputations that increase morbidity and reduce lifespan [1, 2]. A high blood sugar level is responsible for DM, leading to human metabolic disorders. Insulin is a type of hormone released from the pancreas into the bloodstream. Insulin helps glucose enter the body cell from the bloodstream and balance the sugar level. When the pancreas fails to secrete enough insulin, sugar also fails to enter the body cell; subsequently, the sugar level increases and causes diabetes. Diabetes is influenced by various factors such as height, weight, genetic factors, and insulin, but the most important thing is remembering the sugar concentration [3]. It is the cause of many fatal diseases like cardiovascular disease, nerve damage, kidney damage, and depression. They are sorts of type 1 (during childhood), type 2 (at any age), and gestational (pregnant women) [4]. The latest prediction shows that the disease burden of DM had a global prevalence of 425 million people with diabetes in 2017, which is estimated to rise to 629 million by 2045 due to the majority of obesity, physical inactivity, poor diets, sedentary lifestyle, and also genetics [5]. Most of these numerical increments will face in developing countries [6]. According to the world health organization (WHO), more than 77% of patients have reached severe cases due to DM over more than 20 years [7]. Diabetes and its complications affect individuals physically, financially, and socially. According to the report, 1.2 million people die yearly from an untreated health condition. Diabetes-related risk factors, such as cardiovascular and other disorders, resulted in about 2.2 million deaths. Generally, the DM diagnosis process is time-consuming and complex because of the physician’s manipulation. On the other hand, the physician only focuses on the present patient report. But computational detection using a machine learning (ML) algorithm compares the current report with many other factors, which gives a more accurate result. In addition, DM is a life-quality-reducing disease that can lead to more severe issues in the human body. For this reason, it is challenging and essential to diagnose and identify diabetes at the primary phase early. Early diagnosis is a procedure for detecting a disease or disorder in patients in the early stages. It enables people to make important decisions about their care, support, and financial and legal matters. Furthermore, it helps them get crucial information, counsel, and support as they face new problems. However, detecting diabetes early on becomes more challenging due to the uncertainty of the parameters of different physical, environmental, and family backgrounds. Besides, the value of the parameters varies from person to person.
ML is an artificial intelligence- (AI-) based application that automatically builds an analytical model which can be learned from data, identifying the patterns and determining with minimal latency. It can learn something and overcome the deficiencies from experience as humans do [7, 8]. ML-based algorithms are essential for investigating this issue and developing a more accurate CAD scheme for predicting not only the survival rate but also other factors for diabetes in the current era, as they dominate the various tasks of computer vision and the medical industry, including radiology [8, 9]. It is used in the medical field to detect fatal diseases. Also, it assists in streamlining hospital administrative processes, mapping and treating infectious diseases, and personalizing medical treatments [10, 11]. Moreover, ML is also applicable to biological data to extract knowledge by taking the help of feature engineering techniques and diagnosing human-threatening diseases like DM [10]. To accomplish this analysis, random forest is employed with its best parameters. Random forest is a supervised learning method that can be utilized for data classification and prediction. Nonetheless, it is primarily employed to overcome classification issues. The random forest algorithm constructs decision trees from sample data, generates predictions from each one, and then conducts a vote to identify the optimal option. This ensemble technique is preferred to an individual decision tree since it averages the findings to reduce overfitting [11].
Feature engineering converts raw data into features that may be used to construct a prediction model with ML or statistical modeling. It aims to optimize ML models’ performance by preparing an input dataset that best fits the algorithm. In addition, k-fold cross-validation is a frequently employed approach for testing the performance of an ML model. It involves randomly partitioning the data into a collection of folds, where each fold is utilized as a test set in turn while the remaining folds are used as training data. This procedure is done K times, with each fold serving as the test set exactly once. The performance indicator is then averaged over K iterations.
1.2. Motivation
Our research investigated the various aspects of diabetes that helped us identify it early. We used feature engineering techniques to train the algorithm, which helped to provide the best output. In our study, we used a random forest with its best parameters to improve the performance of diabetes identification by employing the tuning parameters and applying the grid search approach. The parameters of RF are tweaked to create a superior classifier that is more robust and precise. GridSearchCV holds all the best parameters to obtain such a type of classifier.
Using the best parameters of a random forest classifier can provide several benefits and could be our study’s novelty and motivating factors. These motivating factors are as follows.
1.2.1. Improved Performance
By tuning the parameters of a random forest classifier, we can often achieve better performance in terms of accuracy, precision, recall, and other evaluation metrics. This is a key motivation for using the best parameters, as they can improve the classifier’s effectiveness [12].
1.2.2. Reduced Overfitting
Overfitting occurs when a model is too complex and has too many parameters, leading to a poor generalization of new data. Using the best parameters of a random forest classifier can reduce the risk of overfitting and improve the model’s ability to generalize to unseen data [13].
1.2.3. Increased Efficiency
Some parameters of a random forest classifier, such as the number of trees and the maximum depth of each tree, can impact the computational efficiency of the model. Using the best parameters, we can optimize the computational efficiency of the classifier and potentially reduce the time and resources required for training and prediction [14].
1.2.4. Enhanced Interpretability
The parameters of a random forest classifier can also affect the interpretability of the model. For example, using the best parameters may result in a simpler and more easily interpretable model, which can also be a motivating factor for using the best parameters in our paper [15].
In summary, using the best parameters of a random forest classifier can lead to improved performance, reduced overfitting, increased efficiency, and enhanced interpretability.
1.3. Contributions
- (i)
We present a random forest algorithm with its optimal parameters (RFWBP) that more effectively diagnoses diabetes in early-stage patients
- (ii)
Our proposed RFWBP achieves much better accuracy when compared with other existing ML algorithms within a short time
- (iii)
We use feature engineering techniques to extract the features from the raw data, taking some preprocessing strategies that help get better performances
- (iv)
We use the k-fold cross-validation technique with the best parameters for the proposed RFWBP algorithm as well as some ML algorithms like decision tree (DT), random forest (RF), support vector machine (SVM), AdaBoost, and linear regression (LR) which determines the detection diabetes, giving the reader better insight regarding the classification approach
The remainder of this work is structured as follows: the literature review discussion is represented by Section 2. Section 3 contains materials and methodologies. Section 4 elaborates on the suggested RFWBP technique. The outcome and discussion are mentioned in Section 5, whereas the conclusion and future recommendations are summarized in Section 6.
2. Literature Review
Diabetes is a chronic and significant health problem that leads to many complications in the human body. Many researchers investigated diabetes using ML techniques to extract features for predicting and identifying diabetes. Sisodia S and Sisodi D [16] proposed predictive analysis models based on DT, SVM, and naive Bayes (NB) algorithms. They got 76.30% as the highest accuracy from NB, which could be improved using a large dataset with some fruitful preprocessing steps. In [17], Alehegn used several ML algorithms, including logistic regression, NB, and SVM, to evaluate the method with 10fold cross-validation [18]. They showed that SVM obtained the best performance and accuracy of 84%. However, the accuracy needed to be increased for the prediction of DM. Perveen et al. [19] looked into the effectiveness of AdaBoost and bagging ensemble ML algorithms in classifying DM patients based on diabetic risk factors utilizing the J48 decision tree as a baseline. The experiment results indicate that AdaBoost surpasses bagging and a J48 decision tree regarding efficiency. Shakeel et al. [4] proposed a cloud-based framework to diagnose DM using k-means clustering, where they compared their work with the other two clustering methods and found better results than the other two. But their framework gives the predicted outcome for only a specific group of affected people. Vijayan and Anjali [3] have taken another ML approach implementing SVM, k-nearest neighbors (KNN), and a decision tree. They found the highest 80.72% accuracy using the AdaBoost algorithm with a decision stamp as a base classifier. Barakat et al. [20] used an SVM classifier to detect DM with good accuracy. Moreover, they used an additional explanation module to make SVM more effective, which helped get better performances. A survey has been done by Shivakumar [21] on data mining technologies for diabetic prediction. After analyzing essential research papers, they found some relation among the diseases like wheezing, edema, oral disease, female pregnancy, and age with having a person diabetes. Choudhury and Gupta [22] surveyed various ML techniques using a dataset (PIMA Indian diabetes dataset) to analyze different models. Finally, they found the best 77.61% of accuracy at LR. SVM and KNN also worked well on that dataset. Sumangali [23] made a model by combining RF and classification and regression tree (CART), which gave them an excellent performance. They also found that a combined classifier model is much more effective than a single classifier model. Experimental work has been done by Chowdhary et al. [24] on diabetes retinopathy detection using ensemble ML algorithms. They found that their model outperformed other existing ML algorithms. Zou et al. [25] tried to detect DM with ML algorithms such as decision trees, random forests, and neural networks. They also used 5-fold cross-validation to examine their model precisely. To reduce the dimensionality, principal component analysis (PCA) and minimum redundancy with maximum relevance have been used and finally found the maximum 80.84% accuracy from the random forest classifier. In [26], Rahman et al. used LR based on p value and odds ratio to predict risk factors for diabetes disease. They proposed a combined LR-based feature selection and RF-based classifier model, which gives better results than other models. Saxena et al. [27] proposed a method using KNN, which acquired an accuracy of 70%, where it should be improved considering a larger dataset. In [28], there is a proposed method based on an NB classifier with good accuracy of 77.01%. In addition, Perveen et al. [19] applied the AdaBoost classifier, offering better performance in detecting DM. However, the work could be more impactful using a large dataset with some preprocessing steps. In [29], Nai-arun and Moungmai used an algorithm to classify the risk of DM. The authors used DT, ANN, LR, and NB ML classification methods to achieve the outcomes. Additionally, bagging and boosting techniques are utilized to increase the consistency of the constructed model. According to the test results, the RF algorithm performed best against all the algorithms used. However, all the associated parameters are needed to fit the model perfectly. Also, they could have increased the performance by using the best parameter of the algorithms.
In prior research, the authors employed traditional statistical machine-learning methods to identify diabetes in tabular data. Their investigation on a few small datasets utilized a black-box-like algorithm that obtained 70-85% accuracy based on their experiment. However, our research employed the random forest technique with its optimal parameters. When using the optimal parameters for an ML algorithm, we are effectively fine-tuning the model to perform optimally on a certain dataset. This can result in enhanced accuracy, precision, and recall, as well as shorter training and inference times, regardless of the dataset size or complexity.
3. Materials and Methods
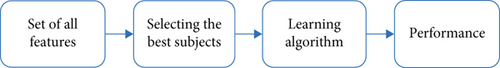
The materials and methods section elaborates the working procedures from first to last, which helps understand the method well-handled. Here, we describe the steps that help to analyze our research study in Figure 1. We use several ML techniques to identify whether the patient has diabetes or not.
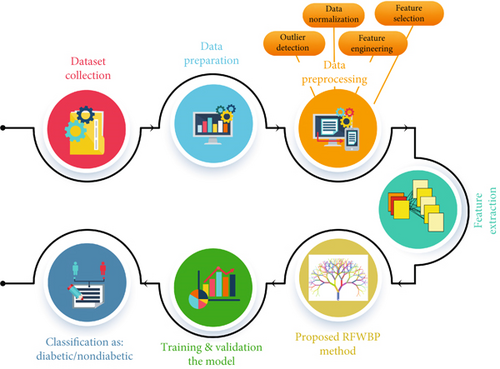
3.1. Dataset
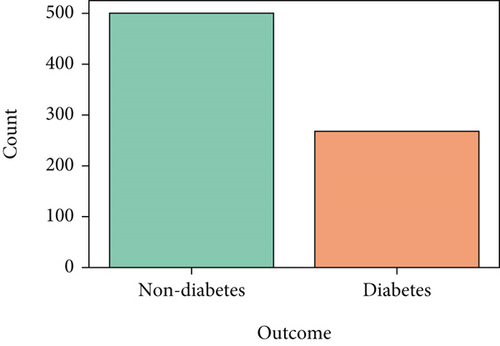
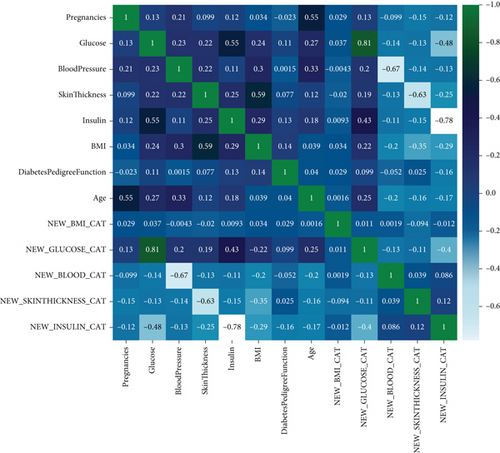
A dataset aggregates some necessary data to help the model perform better. It is fed to the ML algorithm to ensure how accurately the algorithm is interpreted [30]. In our research paper, we used a dataset of different features based on health information to diagnose whether the patient has diabetes. We collected the dataset from Kaggle [31], the world’s largest data science community, with various tools and services to assist in achieving data science objectives. The dataset named “Pima Indians Diabetes Database” contains some health condition features like pregnancies, glucose, blood pressure, age, skin thickness, insulin, BMI, and diabetes pedigree function from the patients, shown in Table 1. The dataset was manipulated by the National Institute of Diabetes and Digestive and Kidney Diseases. The objective of the dataset is to determine the probability that an individual has diabetes based on the specific diagnostic metrics provided in the information. Multiple restrictions governed the inclusion of these occurrences from a more extensive database. There are 768 female patients aged 21-81 years old. The patients’ average age and standard deviation are 33 and 11.76, respectively. Moreover, the descriptive statistics of the dataset are shown in Table 2, explicitly. The numbers of diabetes and no diabetes are 268 and 500, respectively, as shown in Figure 2. Besides this, some parameters have been added (see Table 3) by applying the feature engineering technique further to make this model more precise.
| Pregnancies | Glucose | Blood pressure | Skin thickness | Insulin | BMI | Diabetes pedigree function | Age | Outcome |
|---|---|---|---|---|---|---|---|---|
| 6 | 148 | 72 | 35 | 0 | 33.60000 | 0.62700 | 50 | 1 |
| 1 | 85 | 66 | 29 | 0 | 26.60000 | 0.35100 | 31 | 0 |
| 8 | 183 | 64 | 0 | 0 | 23.30000 | 0.67200 | 32 | 1 |
| 1 | 89 | 66 | 23 | 94 | 28.10000 | 0.16700 | 21 | 0 |
| 0 | 137 | 40 | 35 | 168 | 43.10000 | 2.28800 | 33 | 1 |
| Variable | Distinct | Min | Max | Zeros | Mean | STD | Variance | Skewness | Missing |
|---|---|---|---|---|---|---|---|---|---|
| Pregnancies | 17 | 0 | 17 | 111 | 3.845 | 3.369 | 11.354 | 0.902 | 0 |
| Glucose | 136 | 0 | 199 | 5 | 120.895 | 31.973 | 1022.248 | 0.174 | 0 |
| Blood pressure | 47 | 0 | 122 | 35 | 69.105 | 19.356 | 374.647 | -1.844 | 0 |
| Skin thickness | 51 | 0 | 99 | 227 | 20.536 | 15.952 | 254.473 | 0.109 | 0 |
| Insulin | 186 | 0 | 846 | 374 | 79.799 | 115.244 | 13281.180 | 2.272 | 0 |
| BMI | 248 | 0 | 67.1 | 11 | 31.993 | 7.884 | 62.159 | -0.428 | 0 |
| Diabetes pedigree function | 517 | 0.078 | 2.42 | 0 | 0.472 | 0.331 | 0.109 | 1.919 | 0 |
| Age | 52 | 21 | 81 | 0 | 33.241 | 11.760 | 138.303 | 1.129 | 0 |
| Sex | Female | ||||||||
| Sample size | 768 | ||||||||
| New_BMI_cat | New_glucose_cat | New_blood_cat | New_skin thickness_cat | New_insulin_cat |
|---|---|---|---|---|
| Obese | Prediabetes | Normal | 0 | Abnormal |
| Slightly_fat | Normal | Normal | 0 | Normal |
| Normal | Prediabetes | Normal | 0 | Abnormal |
| Slightly_fat | Normal | Normal | 0 | Normal |
| Obese | Normal | Normal | 0 | Abnormal |
3.2. Data Preprocessing
Preprocessing transforms raw data into form machines, and computers can interpret and evaluate it. Text, photos, video, and other information about the real world are jumbled. In addition to including errors and inconsistencies, it is typically inadequate and needs a consistent style. Since computers prefer to work with clean data, they interpret it as 1 s and 0 s. It is estimated that data preparation accounts for 60% of all effort and time utilized in the data mining process [29–31]. We have utilized some preprocessing strategies in this work, such as data normalization, transformation, outlier identification, feature engineering, and feature selection, detailed in the following subsections.
3.3. Data Normalization
Normalization of data is a preprocessing approach that entails scaling or altering the data to ensure that every attribute contributes equally. The term normalization refers to the process of arranging data into multiple related tables in order to eliminate data redundancy. The performance of ML algorithms depends on the data quality used to build a comprehensive statistical model for the categorization problem. Recent research has highlighted the importance of data normalization for improving data quality and, subsequently, the performance of ML algorithms [32].
3.4. Outlier Detection
where IQR = Q3 − Q1 and k ≥ 0.
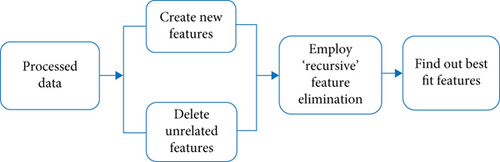
3.5. Feature Engineering
Feature engineering is a significant step before building a precise model. Finding all the necessary features in a compatible format while working with an ML algorithm [44] is crucial. Without these essential features, the algorithm does not perform properly, and the result also goes down. The term feature engineering presents similar activities like improving the existing features (see Figure 3) and adding some new features [45]. It is all about feeding the model and making it more fruitful. Some practical steps of feature engineering include feature generation, feature extraction, feature transformation, feature selection, and feature analysis and evaluation. It is also a method for transforming unprocessed data into features that better address the core problem with ML models, resulting in increased model accuracy on previously unknown data [46]. Our research study has added exclusive features based on existing features of raw data labeled as BMI category, glucose category, blood category, skin-thickness category, and insulin category to get the best performances from our method. Table 4 shows the added features based on the value ranges of raw data.
| New_BMI_cat | New_glucose_cat | New_blood_cat | New_skin thickness_cat | New_insulin_cat | |||||
|---|---|---|---|---|---|---|---|---|---|
| BMI range | BMI label | Glucose range | Glucose label | Blood pressure range | Blood pressure label | Skin thickness range | Skin thickness label | Insulin range | Insulin labels |
| 0-18.4 | Weakness | 0-139 | Normal | 0 - 79 | Normal | 1-18 | Normal | 0-16 | Normal |
| 18.4-25.0 | Normal | 139-200 | Prediabetes | 79-90 | Hypertension_S1 | 19-88 | Abnormal | 17-166 | Abnormal |
| 25.0-30.0 | slightly_fat | — | — | 90-123 | Hypertension_S2 | — | — | — | — |
| 30.0-70 0 | Obese | — | — | — | — | — | — | — | — |
Moreover, one-hot encoding may also use in feature engineering that encodes the categorical variable to a numeric form for the better prediction skill of an ML algorithm [47]. In ML, the dataset contains many categories of data. Some algorithms can work with the categorized values, but most need help. Labeled data is a big problem for them, so the data must be converted into numeric. To make the data more acceptable, we rebuild the encoding dimension of the main network package in the data collection by applying individual-heat coding to generate two-dimensional data. We used a one-hot encoding technique to add a binary variable for each unique categorical value. It deals with only 1 and 0. The actual values are assigned to 1, and the remaining variables are considered false and assigned to 0 [48].
3.6. Feature Selection
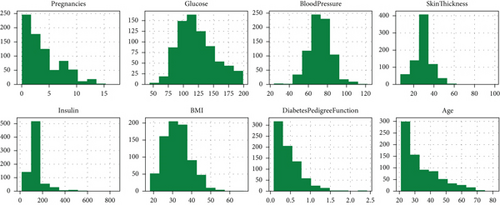
Feature selection automatically or manually selects those features, contributing much to predicting the results from a model [31]. It is classified into three groups based on filters, wrappers, and embeds used for statistical measures between the input variables. The wrapper feature selection method uses an induction learning algorithm to evaluate the feature subset. It measures the performance based on categorizing the rate gained from the testing set. The embedded process uses a particular supervised and nonsupervised ML algorithm to incorporeal sense about the specific form of the class. The filter method shows complete independence between the learning machine and raw data, which is relatively robust against overfitting [49]. They can be filtered to select the relevant features, reducing the noise effects from the overall raw data [50]. It has been discussed in ML and the data mining field to find the best k features and avoid generalization errors in the generalization errors [51]. In addition, Figure 4 depicts the histograms of each feature in our experimented dataset, which is the quickest way to understand the distribution of each attribute in the dataset.
Here, r is the correlation coefficient; xi is the x-variable values in a sample; is the mean of the x-value variables; yi is the y-variable values in a sample; is the mean of the y-value variables [53].
3.7. Feature Extraction
This attribute is very desirable according to the Levinson-Durbin algorithm.
4. Proposed RFWBP Method
Like the RF method, our proposed RFWBP method is a supervised learning technique used for classification and regression problems but primarily used in classification problems. It is blended with the RF algorithm and feature engineering. It selects the best parameters from the total number of parameters and uses them to predict and classify the problem.
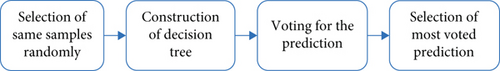
RF with a single tree is a simple decision tree that tends to overfit. The proposed RFWBP algorithm is developed of multiple trees based on the premise that a forest with more trees is more adaptable while reducing model variance. It makes the decision trees on data samples and gives a prediction for each tree to select the solutions by means and voting shown in Figure 7. RFWBP uses exclusive features based on existing raw data features labeled as BMI, glucose, blood, skin thickness, and insulin to get the best performance. The best parameters used in our proposed RFWBP are in Table 5.
| Tuning parameter | Best parameter | Parameter function |
|---|---|---|
| “n_estimators”: [10, 17, 25, 33, 41, 48, 56, 64, 72, 80] | “n_estimators”: 33 | Number of trees that builds before taking the maximum voting |
| “Max_features”: (“auto” and “sqrt”) | “Max_features”: ‘auto’ | Number of features to consider when looking for the best split |
| “Max_depth”: [2, 4] | “Max_depth”: 4 | The depth of each tree in the forest |
| “Min_samples_split”: [2, 5] | “Min_samples_split”: 5 | Minimum number of samples required to split an internal node |
| “Min_samples_leaf”: [1, 2] | “Min_samples_leaf”: 2 | Minimum number of samples required to be at a leaf node |
| “Bootstrap”: (true, false) | “Bootstrap”: true | Involves random sampling of a dataset with replacement |
The training dataset’s cross-validation accuracy and the significance of every element as the performance parameter are measured using the RF algorithm. Fast trees or the basic units of an RF algorithm are distinct and can create collaterally. After that, we choose the best subset by observing the maximum aggregate of the average score and median score with minimum standard deviation (SD). To best prevent the overfitting problem, the k-fold cross-validation technique ensures stable performance.
All the procedures are given below.
Step 1. Using a parallel random forest (PRF) classifier, train the dataset and then measure and sort the median of the variables by their importance through 20 trials
Step 2. Select and add every feature containing the highest variables’ importance and train the dataset by PRF with k-fold cross-validation
Step 3. Compute the score for every feature’s Fi where i = 1 ⋯ n (n expresses the number of features in the executing loop)
Step 4. Choose the best features’ subsets by selecting the rules described below
Step 5. Repeat the steps until it arrives at the expected criteria
- (i)
Rule 1. Choose attributes that have the highest median score
- (ii)
Rule 2. Choose features that have the highest average score
- (iii)
Rule 3. Look for features that have the lowest SD
-
Algorithm 1: Proposed RFWBP method for diabetes identification [53].
-
Input: Final dataset after all the preprocessing steps and feature engineering techniques.
-
1. Use a parallel random forest classifier and sort out the median of variables through 20 trials.
-
2. Train the classifier using PRF with k-fold cross validation and in the jth cross-validation, a set of Fi, is obtained.
-
3. Calculate the Fi score in every feature.
-
4. i = 1 ⋯ ⋯n (n is the number of features).
-
5. (13)
-
6. Choose subsets containing the best features following the rules.
-
(i) Choose attributes that have the highest median score
-
(ii) Choose features that have the highest average score
-
(iii) Look for the features that have the lowest SD
-
7. Repeat the above steps until it arrives at the expected criteria.
-
8. Training with the RFWBP algorithm
-
9. Evaluate the RFWBP model
-
10. Output: result = nondiabetic and diabetic.
The best accuracy and lowest SD are obtained using these guidelines. As a result, the best selection of features tends to minimize the number of output features to the least possible. The RF importance of the component is determined using ML algorithms. We discover the subset of features with negligible characteristics while still accomplishing the problem’s goal based on the estimated relevance value. We have implemented our model using rf_RandomGrid for searching the trees randomly by tuning the parameters that increase the model’s generalizability. Evaluating metrics are used for the conversion from the grid, and random combinations of hyperparameters are considered in every iteration in this search pattern which helps the model to show accurate performance. Furthermore, Algorithm 1 describes the overall Diabetes identification process using the proposed RFWBP method [58].
5. Result and Discussion
Our research study aims to identify diabetic patients based on diabetes risk factors like age, glucose level, blood sugar concentration, pregnancies, BMI, and skin thickness. We evaluated our study on a dataset from Kaggle [31]. The study is implemented using Jupyter Notebook and Google Colab. In this research, we utilized the RFWBP algorithm to achieve the best results when comparing our technique to different ML algorithms, including DT, RF, SVM, NB, and AdaBoost. We also evaluated the findings using a 5-fold cross-validation and an alternative (without cross-validation) based on precision, recall, F1 score, and accuracy.
Throughout the experiment, the performances were divided into three distinct segments. Taking 70% for training with 30% for testing, 80% for training with 20% for testing, and a 5-fold cross-validation technique on the entire dataset. These are as follows:
in Table 6, 70% of the total dataset was utilized for training and 30% for testing. Comparing our proposed RFWBP method (without cross-validation) to existing ML algorithms, we obtained the lowest values from DT and the best deals from RF with the best parameters based on precision, recall, F1 score, and accuracy. Using 5-fold cross-validation, we compared the performance of our proposed method to that of various existing ML algorithms in Table 7. Our proposed classifier attained a maximum accuracy of 95.83% with confidence intervals of 95%. A few ML algorithms performed well; however, NB’s cross-validation technique yielded the worst results. Table 8 contains a performance comparison between our proposed model (without cross-validation) and other existing ML algorithms using 80% of the whole dataset as training data and 20% as testing data based on precision, recall, F1 score, and accuracy. In addition, our suggested model achieved a maximum accuracy of 90.68 percent compared to existing ML methods.
| Model | Precision | Recall | F1 score | Accuracy |
|---|---|---|---|---|
| Decision tree | 92.90 | 91.72 | 92.31 | 88.61 |
| Support vector machine | 91.61 | 89.87 | 90.73 | 87.45 |
| AdaBoost | 94.85 | 88.02 | 91.30 | 87.87 |
| Naive Bayes | 85.81 | 90.48 | 88.08 | 84.42 |
| Logistic regression | 90.83 | 91.67 | 91.24 | 87.66 |
| Random forest | 90.32 | 90.32 | 90.32 | 87.01 |
| Gradient boosting machine | 88.99 | 90.65 | 89.81 | 85.71 |
| CatBoost | 89.91 | 91.59 | 90.74 | 86.84 |
| Multi-layer perceptron | 91.74 | 92.59 | 92.17 | 88.82 |
| Proposed RFWBP | 94.13 | 91.73 | 92.81 | 90.32 |
| Algorithm | 1st fold CV | 2nd fold CV | 3rd fold CV | 4th fold CV | 5th fold CV | Mean CV (accuracy) |
|---|---|---|---|---|---|---|
| Decision tree | 87.01 | 87.66 | 88.31 | 88.24 | 87.58 | 87.76 |
| Support vector machine | 87.66 | 85.72 | 87.01 | 86.28 | 80.40 | 85.42 |
| AdaBoost | 87.66 | 84.42 | 87.01 | 88.24 | 87.58 | 86.98 |
| Naive Bayes | 85.07 | 79.87 | 86.36 | 86.28 | 81.05 | 83.73 |
| Logistic regression | 86.36 | 84.21 | 86.48 | 86.53 | 88.33 | 86.38 |
| Random forest | 87.66 | 84.41 | 87.01 | 89.54 | 88.24 | 87.37 |
| Gradient boosting machine | 88.32 | 87.37 | 88.89 | 91.21 | 89.68 | 89.01 |
| CatBoost | 92.19 | 88.21 | 94.56 | 91.12 | 93.78 | 91.97 |
| Multi-layer perceptron | 91.51 | 91.35 | 95.21 | 92.35 | 94.87 | 93.01 |
| Proposed RFWBP | 95.67 | 95.55 | 95.99 | 96.58 | 95.35 | 95.83 |
| Model | Precision | Recall | F1 score | Accuracy |
|---|---|---|---|---|
| Decision tree | 88.99 | 91.51 | 90.23 | 86.36 |
| Support vector machine | 89.91 | 90.74 | 90.32 | 86.36 |
| Logistic regression | 90.83 | 91.67 | 91.24 | 87.66 |
| AdaBoost | 90.83 | 88.39 | 89.59 | 85.06 |
| Naive Bayes | 83.49 | 90.10 | 86.67 | 81.82 |
| Random forest | 92.66 | 90.99 | 91.82 | 88.31 |
| Gradient boosting machine | 88.07 | 91.43 | 89.72 | 85.53 |
| CatBoost | 88.99 | 93.27 | 91.08 | 87.50 |
| Multi-layer perceptron | 91.74 | 93.46 | 92.59 | 89.47 |
| Proposed RFWBP | 92.38 | 94.21 | 93.29 | 90.68 |
In Table 9, we compared the results of our proposed RFWBP approach to the existing related work that they obtained from their research. It implies that our proposed method employing the best RF parameters provided the best results.
| Authors | Algorithm | Accuracy (%) | Year |
|---|---|---|---|
| Saxena et al. [27] | KNN | 70.00 | 2014 |
| Rani and Jyothi [28] | NB | 77.01 | 2016 |
| Choudhury and Gupta [22] | LR | 77.61 | 2019 |
| Vijayan and Anjali [3] | AdaBoost | 80.72 | 2015 |
| Zou et al. [25] | RF | 80.84 | 2018 |
| Faruque [18] | SVM | 84.00 | 2019 |
| Khanam and Foo [59] | ANN | 88.60 | 2021 |
| Proposed method | RFWBP | 95.83 | 2023 |
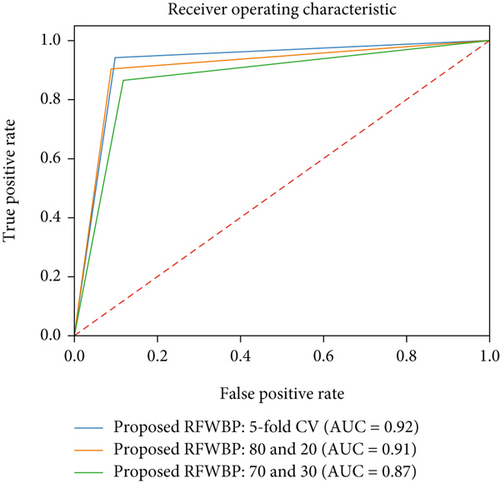
Figure 8 shows the graphical representation of the performance of our proposed classifier. It plots the true positive rate on the y-axis against the false positive rate on the x-axis at different classification thresholds. In creating a ROC curve, the classifier is first trained on a dataset and then tested on a separate dataset. The true positive and false positive rates for each classification threshold are calculated and plotted on the ROC curve. The resulting curve shows the trade-off between the true positive rate and the false positive rate for the classifier. Observing the curve, we find that the area under the curve (AUC) is 0.92, while we used 5-fold cross-validation on our proposed RFWBP classifier. A ROC curve can also be used to compare different classifiers’ performance and identify the optimal classification threshold for a given classifier. Furthermore, the mean squared error (MSE) determined by the Python function is 0.0117.
Regarding activity versus better performance, gathering more data and feature engineering pays off the most. Still, once we have saturated all databases, it is time to move on to model hyper-parameter tuning. Random forest parameters are often used to boost the model’s prediction power or make it easier to train. Hyperparameters are best compared to the settings of an algorithm that can be tweaked to improve performance.
In our study, we used the best parameters of the random forest algorithm instead of default parameters. Hence, we got the best performances that the random forest algorithm shows with its default parameters. Our dataset needs to have some best features from which we may get the best performances. We evaluated the dataset with the default parameter of the random forest, but it shows fewer performances than the random forest with the best parameters. A detailed discussion of our proposed method is conducted based on numerical performance and visual results. In this study, we got a reasonable identification rate with the considerable help of data processing techniques described in the preprocessing section. After several times of fine-tuning, we got the best results using RF with its best parameters as a classifier. In our study, the RFWBP model was exclusively applied to data collected from Pima Indians. We will examine the performance of our suggested approach on other large datasets in the near future. Besides, k-fold cross-validation reduces the variation of the performance estimate by averaging the performance of multiple test sets. In addition, it enables us to use all the data for training and testing in order to obtain a more accurate estimate of the model’s performance, which is essential when data is scarce. Furthermore, as it requires fewer iterations than other validation techniques, such as leave-one-out cross-validation, it is more computationally efficient. The performance estimate can be used to determine the hyperparameters that result in the best model performance.
The proposed method can be deployed in a computer-aided diagnosis system that will help effectively to identify diabetic patients at the early stage. In addition, the early identification of diabetes growth in humans, especially those without admittance to doctors, can significantly encourage them to get the treatment and enrich the survival possibility.
6. Conclusion and Future Work
In this study, we propose a method using the random forest algorithm with its best parameters to assemble a comprehensive data set, including diabetic and nondiabetic patients, to figure out the issue of inaccurate-accurate conclusions in diabetes identification. A medical diagnosis requires lots of information on the patient’s physical condition. The motive for using these parameters was the same. It can detect the abnormality and identify the diabetic patient quickly in a short time. We have shown how to identify diabetes in two ways in our study. Finally, we got 95.83% of the highest accuracy using 5-fold cross-validation and 90.68% accuracy without k-fold cross-validation. Experimental results implied better accuracy, and the mentioned procedure has identical to other diabetes detection algorithms. When applied clinically, our proposed method can be used to detect diabetes quite accurately and precisely. Additionally, it will aid any organization’s ability to diagnose many diabetes patients. However, it has some risk factors, such as incorrect blood glucose and insulin information, which reduces the ability to diagnose diabetes. The number of samples in our study is modest, and the results may need to be more generalizable to other groups or contexts due to the sample. The results of this study might not apply to real-world situations due to its artificial character or controlled conditions. In the future, we will extend our analysis by maximizing the number of subjects and features of both balanced and imbalanced datasets, which could provide detailed insights into the aspects that allow our model to identify diabetes patients more precisely.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Authors’ Contributions
Shahin Ali contributed to the conceptualization, methodology, software, visualization, writing—original draft preparation, and reviewing and editing. Khairul Islam contributed to the supervision, and writing—reviewing and editing. A Arjan Das contributed to the data curation, validation, and writing. D U S Duranta contributed to the data curation and writing. Farija Haque contributed to the data curation and writing. Md Habibur Rahman was responsible for supervision, and writing-reviewing and editing.
Acknowledgments
This work was supported by the Department of Biomedical Engineering (BME), Islamic University, Kushtia 7003, Bangladesh.
Open Research
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.




