Development and Validation of a New Analytical HPLC-PDA Method for Simultaneous Determination of Cucurbitacins B and D from the Roots of Trichosanthes kirilowii
Abstract
Trichosanthes kirilowii, one of the herbal formulas named SH003, has been used for treatment in traditional medicine. This paper aimed to analyze the marker compounds from the roots of T. kirilowii and evaluate a validation method for cucurbitacins B and D using high-performance liquid chromatography-photodiode array detector (HPLC-PDA). Two marker compounds were identified as cucurbitacin B (1) and cucurbitacin D (2) from the roots of T. kirilowii by spectroscopic analyses. Cucurbitacins B and D peaks were well separated and were detected at 15.4 min and 12.4 min, respectively, by a UV detector at 230 nm with the linearity (R2 > 0.999) range between 5 and 250 μg/mL, limit of detections (LODs) were 1.87 μg/mL and 1.30 μg/mL, respectively, while limit of quantifications (LOQs) were 5.66 μg/mL and 3.93 μg/mL, respectively. The established method offered good precision with overall intra- and inter-day variations of 0.34–1.26 and 0.26–1.35%, respectively, for % relative standard deviation (RSD, acceptance limit %RSD <3%). The cucurbitacins B and D recoveries ranged from 99.2 to 101.7% and 98.6 to 102.0%, respectively. These results suggest that cucurbitacins B and D could be a valuable candidate for marker compounds of the extract from T. kirilowii, and the proposed method was shown applicability for quality control of SH003.
1. Introduction
Trichosanthes kirilowii is a perennial vine of the Cucurbitaceae family. Its roots and seeds have been used in traditional medicine as an anti-inflammatory agent, cough, phlegm, polydipsia, diabetes, and sore throat medicine [1, 2]. Previous phytochemical investigations of T. kirilowii have yielded lignans, flavones, and triterpenes [3, 4]. T. kirilowii is one of the herbal formulas named SH003, which is known by the Korean common name of hanultari. SH003 is a mixed herbal extract based on the principle of traditional medicine containing Astragalus membranaceus, Angelica gigas, and Trichosanthes kirilowii [5]. Each of these herbs has long been used in traditional medicine and several anticancer effects have been reported [6–10]. SH003 has been reported to show various anticancer effects such as breast, lung, prostate, cervical, and gastric cancer through several in vitro and in vivo studies [11–15]. As SH003 is an herbal medicine composed of numerous compounds, the quality is based on the analysis of one or more selected compounds. A characterization of SH003 was based on the retention times and UV spectra of standard chemicals such as decursin and nodakenin from A. gigas and formononetin from A. membranaceus. However, the index compound for T. kirilowii [16] was not detected in the previous analytical method [5]. Zhang H et al. reported the mass spectrometry coupled with the UHPLC method for the simultaneous determination of two categories of ingredients from different parts of T. kirilowii, however, excessively high sensitivity is a disadvantage of HPLC-MS, which is not conducive to the detection of marker compounds in complex systems of traditional Chinese medicine. Therefore, in order to demonstrate the properties of SH003 as a novel herbal medicine, such as anticancer treatment, a systematic investigation using HPLC-PDA is more suitable for the development of new marker components from the roots of T. kirilowii.
The present study aims to isolate the secondary metabolites from the roots of T. kirilowii and validate a simple, rapid, and economical method using HPLC-PDA for the analysis of marker compounds, which has not been addressed.
2. Materials and Methods
2.1. General Experimental Procedure
1H and 13C-NMR spectra were recorded on a Bruker 800 MHz spectrometer for 1H and 200 MHz for 13C, respectively. 1H–1H COSY, HMBC, and HMQC spectra were obtained with the usual pulse sequences and data processing was performed with standard software. Chemical shifts are given in ppm referring to the CD3OD signal set at δH 3.33 and 4.88 or middle band set at δC 49.0. TLC and column chromatography were carried out on precoated silica gel F254 Plate (Merck, art. 5715), RP-18 F254S Plate (Merck, art. 15389), silica gel 60 (230–400 mesh, Merck), Sephadex LH-20 (bead size 25–100 μm, Sigma), and Toyopearl HW-40C (50–100 μm, Tosoh).
2.2. Reagents and Chemicals
HPLC grade methanol and acetonitrile were purchased from J.T. Baker Co. (Phillipsburg, NJ, USA), and deionized water was obtained from a Milli-Q water purification system (Millipore, Billerica, MA, USA). The other reagents and chemicals were purchased from different commercial suppliers and were of analytical grade.
2.3. Plant Materials
The root of T. kirilowii was purchased from Puremind Co, Gyeongsangbuk-do, Korea, on September 2021 and identified by Prof. Ho Young Choi, College of Oriental Medicine, Kyung Hee University. The voucher specimen (822–15) was deposited at the herbarium of the Korea Institute of Science and Technology (KIST).
2.4. Extraction and Isolation
The roots of T. kirilowii (30 g) were extracted with H2O/EtOH (30% v/v, 150 mL) three times at 85°C for 3 h. The extracts were filtered through celite and evaporated under reduced pressure to yield brown powder. These hydroethanolic extracts (10.1 g) were dissolved in MeOH (100 mL) and then ultrasonication was performed at room temperature for 30 min. The experiments were carried out in three replicates. After filtration, the MeOH soluble fraction (2.4 g) was fractionated on a Sephadex LH-20 chromatography and eluted with MeOH to yield eight fractions (MS1–MS8). Fraction MS4 (726.6.mg) was subjected to Toyopearl HW-40 column eluting with MeOH to give eight fractions (MS4a–MS4h). Fraction MS4b (357.4 mg) was purified using a RP-18 column eluted with a gradient mobile phase system (MeOH : H2O, 30 : 70 to 100 : 0), and purified further by preparative reverse-phase HPLC using a Luna C18 (5 μm, 250 × 10 mm, phenomenex, Torrance, CA, USA) eluted with 40% ACN, yielded 1 (1.1 mg) and 2 (3.0 mg).
2.4.1. Cucurbitacin B (1)
1H-NMR (CD3OD, 800 MHz): δ 0.89 (s, 3H, H-18), 1.05 (s, 3H, H-19), 1.22 (d, J = 12.8 Hz, H-1), 1.30 (s, 3H, H-28), 1.31 (s, 3H, H-29), 1.40 (s, 3H, H-21), 1.42 (s, 3H, H-30), 1.46 (m, H-15), 1.56 (s, 3H, H-26), 1.59 (s, 3H, H-27), 1.87 (dd, J = 9.2, 13.2 Hz, H-15), 1.99 (d, J = 8.0 Hz, H-8), 2.02 (s, CH3CO), 2.05 (m, H-7), 2.11 (m, H-1), 2.43 (dd, J = 8.0, 19.2 Hz, H-7), 2.56 (d, J = 15.2 Hz, H-12), 2.60 (d, J = 7.2 Hz, H-17), 3.00 (br d, J = 12.8 Hz, H-10), 3.42 (d, J = 14.4 Hz, H-12), 4.55 (m, H-16), 4.59 (m, H-2), 5.83 (m, H-6), 6.86 (d, J = 16.0 Hz, H-23), 6.99 (d, J = 16.0 Hz, H-24); 13C-NMR (CD3OD, 200 MHz) : Table 1.
| C | 1 | 2 | C | 1 | 2 |
|---|---|---|---|---|---|
| 1 | 35.7 | 35.7 | 17 | 58.1 | 58.6 |
| 2 | 71.4 | 71.4 | 18 | 19.3 | 19.3 |
| 3 | 212.5 | 212.5 | 19 | 18.3 | 18.7 |
| 4 | 50.4 | 50.3 | 20 | 78.5 | 78.8 |
| 5 | 140.6 | 140.7 | 21 | 24.0 | 23.9 |
| 6 | 119.8 | 119.8 | 22 | 203.5 | 203.8 |
| 7 | 23.4 | 23.4 | 23 | 119.8 | 121.2 |
| 8 | 42.7 | 42.7 | 24 | 153.9 | 150.1 |
| 9 | 47.9 | 48.2 | 25 | 70.2 | 79.6 |
| 10 | 33.4 | 33.4 | 26 | 28.4 | 25.5 |
| 11 | 214.2 | 214.4 | 27 | 27.8 | 25.0 |
| 12 | 48.4 | 48.4 | 28 | 20.4 | 20.4 |
| 13 | 47.9 | 48.0 | 29 | 27.7 | 28.4 |
| 14 | 50.4 | 50.4 | 30 | 18.1 | 18.0 |
| 15 | 45.2 | 45.0 | CH3CO | 20.4 | |
| 16 | 70.1 | 70.5 | CH3CO | 170.5 |
- ∗Shift values (δ) are expressed in ppm.
2.4.2. Cucurbitacin D (2)
1H-NMR (CD3OD, 800 MHz): δ 0.94 (s, 3H, H-18), 1.06 (s, 3H, H-19), 1.23 (d, J = 12.8 Hz, H-1), 1.30 (s, 3H, H-28), 1.32 (s, 3H, H-29), 1.34 (s, 3H, H-21), 1.35 (s, 3H, H-30), 1.39 (m, H-15), 1.41 (s, 3H, H-26), 1.42 (s, 3H, H-27), 1.87 (dd, J = 9.6, 12.8 Hz, H-15), 1.99 (d, J = 8.0 Hz, H-8), 2.04 (dd, J = 5.8, 19.2 Hz, H-7), 2.11 (m, H-1), 2.44 (dd, J = 8.0, 19.2 Hz, H-7), 2.62 (m, H-17), 2.63 (m, H-12), 3.00 (br d, J = 12.8 Hz, H-10), 3.45 (d, J = 14.4 Hz, H-12), 4.50 (t, J = 8.0 Hz, H-16), 4.58 (dd, J = 4.8, 12.8 Hz, H-2), 5.83 (dd, J = 1.6, 4.0 Hz, H-6), 6.85 (d, J = 15.2 Hz, H-23), 6.99 (d, J = 15.2 Hz, H-24); 13C-NMR (CD3OD, 200 MHz) : Table 1.
2.5. HPLC Analysis
HPLC was performed on Waters 1500 series HPLC system (Waters) equipped with a 1525 binary pump, column oven, and photodiode array detector (model 2996) using a Luna C18 column (5 μm, 250 × 4.6 mm, phenomenex, Torrance, CA, USA). The gradient mobile phase consisted of acetonitrile (ACN) and water (H2O), and the operating conditions are shown in Table 2.
| Time (min) | % ACN | % DW |
|---|---|---|
| Initial | 15 | 85 |
| 10 | 60 | 40 |
| 20 | 80 | 20 |
| 30 | 100 | 0 |
| 35 | 15 | 85 |
The temperature of the column was maintained at 30°C during the chromatographic separation. The sample and solvent were filtered through 0.45 μm PVDF syringe filter and membrane filter and were degassed using an ultrasonic bath prior to use. The flow rate was set at 1.0 mL/min for 35 minutes in a gradient mode. The identification of the chromatographic peaks was performed by comparing the retention times of the samples by recording the UV spectra of the peaks in the range of 200–400 nm.
2.5.1. Preparation of Standard Solution
A stock solution of cucurbitacins B and D was prepared at a concentration of 1000 μg/mL, respectively. One milligram of cucurbitacins B and D was accurately weighed and transferred into a 1 mL volumetric flask and 500 μL of the methanol was added and sonicated for 10 min, the final volume was made up to 1 mL using methanol to produce a final concentration of 1 mg/mL, respectively. The working solution for calibration use was diluted with MeOH to a series of concentrations.
2.5.2. Preparation of Sample Solutions
The powder of 30% EtOH extract from the root of T. kirilowii (100.0 mg) was accurately weighed and transferred into a 10 mL volumetric flask and 5 mL of methanol was added and sonicated for 10 min, and the final volume was made up to the mark of the flask using methanol followed by 5 min shaking. This solution was filtered, and the filtrate was stored at 4°C and filtered through a 0.45 μm membrane filter before analysis.
2.5.3. Method Validation
The linearity was obtained by preparing a series of concentrations of stock solution with at least six appropriate concentrations in duplicate. The validation of this method for the analysis of cucurbitacins B and D was examined in accordance with the guidelines of the International Conference on Harmonization (ICH) by evaluating linearity, precision (intra- and inter-day), accuracy, limit of detection (LOD), and limit of quantification (LOQ)[17].
2.5.4. Specificity and Selectivity
Specificity and selectivity of the analytical method were analyzed, the extent of any interference caused by the blank sample in the retention of the analyte, which were assessed by preparing and examining a blank sample and standard solutions. The absence of detectable interfering peaks at the retention time of the analyte was considered as a lack of interference.
2.5.5. Linearity and Range
The stock solutions of cucurbitacins B and D were diluted in the concentration range of (5–250 μg/mL) in triplicate (n = 3). The linearity curve was prepared by plotting the area of peak obtained versus concentration of the cucurbitacins B and D, respectively. Least-squares linear regression analysis using Microsoft Excel 2016 and Waters Millennium Empower 3.0 software were used to determine the slope, intercept, and correlation coefficient values.
2.5.6. LOD and LOQ
2.5.7. Precision, Accuracy, and Recovery
Precision (intra- and inter-day) of the proposed method was performed on three replicate sets of three concentration samples of cucurbitacins B and D (250, 100, and 50 μg/mL each). Intra-day precision was assessed by injecting 3 independent combined samples on the same day, and inter-day precision was injected using same concentration solutions by comparing the results on 3 different days under the same operating conditions for reproducibility. The precision of the method was expressed as the % relative standard deviation (RSD), and values of %RSD within 3% were acceptable.
3. Results and Discussion
3.1. Structural Elucidation of Compounds 1 and 2
The structures of isolated compounds (Figure 1) were identified on the basis of spectral data (1H-NMR, 13C-NMR, and 2D NMR including H–H COSY, HSQC, and HMBC) (Supplementary material) and by comparison with those reported literature data.
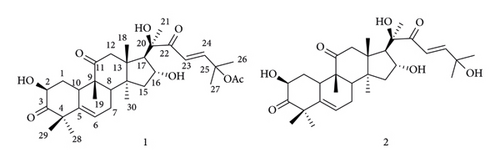
Compound 1 was obtained as a white amorphous solid. The 1H-NMR spectrum showed a pair of doublets for the trans-olefinic protons, oxymethine signals, as well as the signals of eight methyl and a sharp characteristic singlet assigned to an acetate group and accommodated at C-25 hydroxyl groups. 13C-NMR shifts (Table 1) and 2D NMR spectra showed three carbonyl and three oxygenated carbons, two transcoupled olefinic function, one acetyl, and eight methyl groups. This indicated that compound 1 is cucurbitacin B, which was proven through comparison of previously published data [4, 18].
Compound 2 was obtained as a white amorphous solid. The 1H and 13C-NMR spectra showed similarity to those of cucurbitacin D (1), except for the absence of an acetyl group, which was also confirmed by 13C-NMR shifts (Table 1) and 2D NMR. On the basis of the above spectroscopic data, compound 2 was identical to that of the previously known cucurbitacin D [4, 18].
3.2. Method Development
The mobile phase condition in HPLC was also studied, indicating that the gradient eluting system of acetonitrile-water mixture (Table 2) provided excellent separation in a reasonable time.
3.3. Specificity and Selectivity
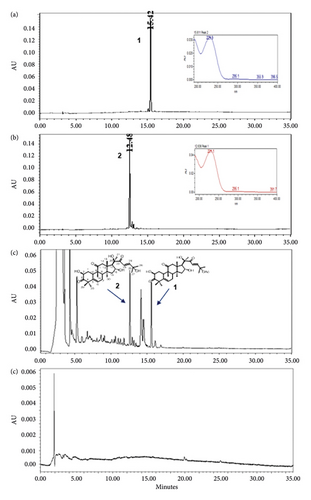
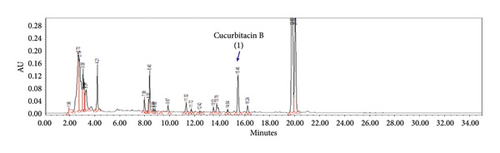
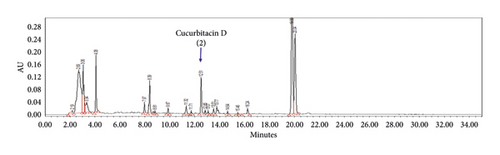
The optimized analytical method was able to detect and assess cucurbitacins B and D in the presence of matrix components. Chromatographic specificity of the method was demonstrated by the absence of significant interfering peaks at the retention time of cucurbitacins B (Rt 15.4 min) and D (Rt 12.4 min) on comparison chromatograms of blank with those of standard as shown in Figure 2. The obtained result indicates that the developed method was specific for cucurbitacins B and D.
3.4. Selection of UV Wavelength
Cucurbitacins B and D have a λmax at 229 and 231 nm, respectively, in a solvent system of water and acetonitrile mixture. An acceptable response was obtained upon the detection of both cucurbitacins at 230 nm either individually (a) and (b) or in combination (c) with retention time (Rt) being achieved at 15.4 min for cucurbitacin B and 12.4 min for cucurbitacin D, as shown in Figure 2.
Figure 3 shows the chromatograms of the mixed herbal extract (a) of the proposed method for quality control of SH003, and spiked with cucurbitacins B (b) and D (c) from T. kirilowii, respectively. Furthermore, it was shown that the peaks of cucurbitacins B and D are well represented without interference from other peaks of components from Astragalus membranaceus and Angelica gigas in the profile of SH003.
3.5. Method Validation
3.5.1. Linearity and Sensitivity
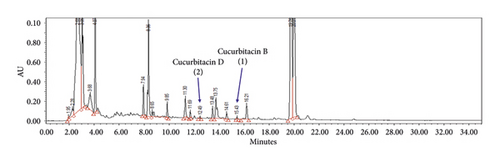
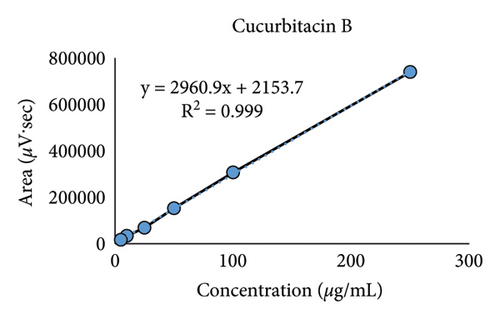
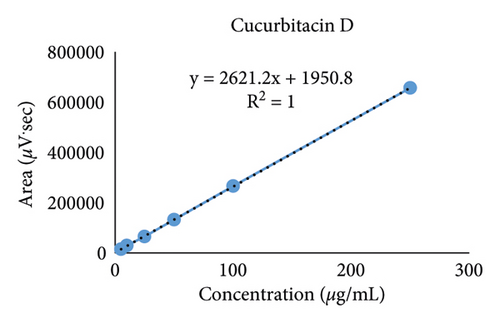
The calibration curves were achieved from 6 different concentrations of cucurbitacins B and D ranging from 5.0 to 250 μg/mL as shown in Figure 4. A high correlation coefficient (R2 > 0.999) value for each calibration curve indicated excellent linearity in this study (Table 3).
| Sample | Retention time (min) | Linear regression equation | Correlation coefficient (R2) | LOD | LOQ |
|---|---|---|---|---|---|
| (μg/mL) | |||||
| Cucurbitacin B | 15.450 ± 0.042 | y = 2960.9x + 2153.7 | 0.999 | 1.87 | 5.66 |
| Cucurbitacin D | 12.589 ± 0.046 | y = 2621.2x + 1950.8 | 1 | 1.30 | 3.93 |
3.5.2. LOD and LOQ
Under our analytical conditions, the LODs of cucurbitacins B and D were determined to be 1.87 and 1.30 μg/mL while the LOQs were 5.66 and 3.93 μg/mL, respectively (Table 3), which reflect the sensitivity of the developed method.
3.5.3. Accuracy, Precision, and Recovery
The inter- and intra-day precisions were determined using standard solution at three concentrations (low, middle, and high quality controls). The intra- and inter-day accuracy and precision were in agreement with the acceptance criteria, and the results were summarized in Table 4. The intra-day RSD values of the cucurbitacins B and D were 0.82–1.26 and 0.34–0.98%, while the inter-day RSD values were 0.74–1.31 and 0.26–1.35%, respectively. All three levels in the % RSD values that are well below 3%, these results showed excellent precision. The results of percentage RSD are also within the prescribed limits as per Q2 (R1) guideline [17].
| Sample | Conc. (µg/mL) | Intra-day ∗ | Inter-day ∗ | ||
|---|---|---|---|---|---|
| Observed conc. (µg/mL) | RSD (%) | Observed conc. (µg/mL) | RSD (%) | ||
| Cucurbitacin B | 50 | 50.3 ± 0.60 | 1.19 | 51.7 ± 0.38 | 0.74 |
| 100 | 103.4 ± 0.85 | 0.82 | 103.2 ± 1.13 | 1.09 | |
| 250 | 248.9 ± 3.14 | 1.26 | 249.0 ± 3.27 | 1.31 | |
| Cucurbitacin D | 50 | 50.9 ± 0.50 | 0.98 | 51.0 ± 0.69 | 1.35 |
| 100 | 100.8 ± 0.34 | 0.34 | 100.1 ± 0.61 | 0.61 | |
| 250 | 249.7 ± 1.90 | 0.76 | 247.9 ± 0.64 | 0.26 | |
- ∗Intra- and inter-day: three times per day and two times analysis of cucurbitacins B and D for three days, respectively.
Table 5 shows the absolute recoveries of cucurbitacins B and D from hydroethanolic extract after spiking different amounts of the standards. The recoveries were in the range of 99.2 ± 1.30% to 101.7 ± 0.61% for cucurbitacin B and 98.6 ± 1.47% to 102.0 ± 0.41% for cucurbitacin D. The recovery values are within the acceptance limit of 98.6–102.0%, indicating good recovery of replicate injections on the HPLC system.
| Compound | Spiked amount (μg/mL) | Measured amount (μg/mL) | Recovery (%)a | RSD (%) |
|---|---|---|---|---|
| Cucurbitacin B | 25.0 | 25.4 ± 0.15 | 101.7 ± 0.61 | 0.60 |
| 50.0 | 49.6 ± 0.65 | 99.2 ± 1.30 | 1.31 | |
| 75.0 | 75.7 ± 0.08 | 101.0 ± 0.11 | 0.11 | |
| Cucurbitacin D | 25.0 | 25.5 ± 0.10 | 102.0 ± 0.41 | 0.40 |
| 50.0 | 49.3 ± 0.74 | 98.6 ± 1.47 | 1.49 | |
| 75.0 | 75.6 ± 0.61 | 100.8 ± 0.82 | 0.81 | |
- aRecovery (%) = (found amount−original amount)/spiked amount ×100.
3.5.4. Quantitative Analysis of Cucurbitacins in T. kirilowii
The proposed validated HPLC method was used to determine the amount of cucurbitacins B and D in the hydroethanolic extract from the root of T. kirilowii. The peak areas of triplicate samples were analyzed by the regression equation obtained from calibration curves to get the content of cucurbitacins B and D. Table 6 shows the contents of cucurbitacins B and D with 0.86 ± 0.01 mg/g and 1.03 ± 0.01 mg/g, respectively. The RSD (%) of cucurbitacins B and D was found to be 0.74% and 1.18%, respectively.
| Cucurbitacins | Contents (mg/g) ∗ | RSD (%) | % (w/w) |
|---|---|---|---|
| Cucurbitacin B | 0.86 ± 0.01 | 0.74 | 0.09 |
| Cucurbitacin D | 1.03 ± 0.01 | 1.18 | 0.10 |
- ∗Values are mean ± SD in triplicate (n = 3).
Several studies have been published to point out that cucurbitacin derivatives exhibit strong anticancer activities against liver, lung, breast, melanoma, and pancreatic cancer [19–22]. These cucurbitacins were known to be widely distributed in the Cucurbitaceae family, even though in low content, cucurbitacins B and D were the main principle components of the hydroethanolic extract from the roots of T. kirilowii.
4. Conclusions
In conclusion, this study utilized a simple simultaneous validation by reversed-phase HPLC with PDA, to accurately quantify cucurbitacins B and D content in T. kirilowii along with medicinal herb sources such as SH003. It was experienced that the proposed analytical method showed to fulfill the requirements for system suitability. HPLC analytical method has been validated by confirming the specificity, linearity, precision, and accuracy to be within the MFDS guidelines. The method could be a useful tool for the separation and determination of cucurbitacins B and D from T. kirilowii, thereby it may be the optimum results for standardization. These results also suggest that cucurbitacins B and D could be a valuable candidate for marker compounds of the extract from the root of T. kirilowii as well as SH003. This report provides an analytical method (HPLC-PDA). This new established method would bring significant benefits for evaluating the quality of T. kirilowii.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors’ Contributions
HJK and SGK conceptualized, supervised, validated, visualized, and investigated the study, performed data curation, formal analysis, funding acquisition, and project administration, collected resources, and developed software. HJK and JYL wrote and reviewed the article. CC and SGK performed the methodology. All authors have read and agreed to the published version of the manuscript.
Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2020R1A5A2019413). The authors thank Prof. Ho Young Choi, College of Oriental Medicine, Kyung Hee University, for the identification of the plant material.
Open Research
Data Availability
The data used to support the findings of this research study are included within the article. However, further data may be obtained from the corresponding author upon reasonable request.