An Improved Differential Evolution Algorithm Based on Dual-Strategy
Abstract
In recent years, Differential Evolution (DE) has shown excellent performance in solving optimization problems over continuous space and has been widely used in many fields of science and engineering. How to avoid the local optimal solution and how to improve the convergence performance of DE are hotpot problems for many researchers. In this paper, an improved differential evolution algorithm based on dual-strategy (DSIDE) is proposed. The DSIDE algorithm has two strategies. (1) An enhanced mutation strategy based on “DE/rand/1,” which takes into account the influence of reference individuals on mutation and has strong global exploration and convergence ability. (2) A novel adaptive strategy for scaling factor and crossover probability based on fitness value has a positive impact on population diversity. The DSIDE algorithm is verified with other seven state-of-the-art DE variants under 30 benchmark functions. Furthermore, Wilcoxon sign rank-sum test, Friedman test, and Kruskal–Wallis test are utilized to analyze the results. The experiment results show that the proposed DSIDE algorithm can significantly improve the global optimization performance.
1. Introduction
Differential Evolution (DE) is an emerging optimization technique proposed by Storn and Price [1] in 1995, which was initially used to solve Chebyshev polynomials. Later, it is demonstrated that DE is also an effective method to solve complex optimization problems. Similar to other intelligent evolutionary algorithms, DE is a stochastic parallel optimization algorithm based on swarm intelligence, which guides optimization search by imitating heuristic swarm intelligence generated by cooperation and competition among individuals in the population.
In DE, the population consists of several individuals, each of which representing a potential solution to an optimization problem. DE generates offspring individuals through mutation, crossover, and selection, and the offspring individuals are expected to be closer to the optimal solution. In the process of evolution, with the increase of generations, the population diversity becomes worse, leading to premature convergence or evolutionary stagnation, which is undoubtedly fatal to the algorithm that depends on the difference of population. Also, the performance of DE is affected by control parameters [2, 3]. For different optimization problems, these control parameters often need a large number of repeated experiments to adjust to the appropriate value for achieving better optimization effect.
To address these shortcomings in DE, many improvements have been proposed, most of which focused on control parameters and mutation strategies.
Population size NP, scaling factor F, and crossover probability CR are three crucial control parameters in DE. Experiments in many works of literatures show that the performance of DE can be improved by adjusting these control parameters. Omran et al. [4] proposed a self-adaptation scheme (SDE), in which F was adaptive and CR was generated by a normal distribution. Liu and Lampinen [5] proposed a fuzzy adaptive differential evolution algorithm (FADE), which used the fuzzy logic controller to adjust F, and CR dynamically and successfully evolved individuals and their fitness values as input parameters of the logic controller. Brest et al. [6] developed a new adaptive DE algorithm, named jDE, applying F and CR to the individual level. If a better individual is produced, these parameters would be retained; otherwise, they would be adjusted according to two constants. Noman et al. [7] proposed an adaptive differential evolution algorithm (aDE), which was similar to jDE [6], except that the updating of parameters in aDE depended on whether the offspring was better than the average individual in the parent population. Asafuddoula et al. [8] used roulette to select the suitable CR value for each individual in each generation of the population. Tanabe and Fukunaga [9] proposed the success-history-based parameter adaptation for differential evolution (SHADE), which generated new F and CR pairs by sampling the nearby space of stored parameter pairs. Later, they came up with an improved version called L-SHADE [10]. Based on SHADE, a linear population size reduction strategy (LPSR) was adopted to reduce the population size NP by a linear function continuously. Zhu et al. [11] proposed an adaptive population tuning scheme (APTS) that dynamically adjusted the population size, in which redundant individuals were removed from the population or “excellent” individuals were generated. Zhao et al. [12] proposed a self-adaptive DE with population adjustment scheme (SAPA) to tune the size of the offspring population, which contained two kinds of population adjustment schemes. Pan et al. [13] proposed a parameter adaptive DE algorithm on real-parameter optimization, in which better control parameters F and CR are more likely to survive and produce good offspring. An enhancing DE with novel parameter control, referred to as DE-NPC, was proposed by Meng et al. [14]. The update of F and CR was based on the location information of the population and the success probability of CR, respectively, and a combined parabolic-linear population size reduction scheme was adopted. Di Carlo et al. [15] proposed a multipopulation adaptive version of inflationary DE algorithm (MP-AIDEA), the parameters F and CR of which were adjusted together with the local restart bubble size and the number of local restarts of Monotonic Basin Hopping [16]. Li et al. [17] presented an enhanced adaptive differential evolution algorithm (EJADE), in which CR sorting mechanism and dynamic population reduction strategy were introduced.
To improve the optimization performance and balance the contradiction between global exploration and local exploitation, researchers have carried out a lot of work on mutation strategy in DE. Das et al. [18] proposed an improved algorithm based on “DE/current-to-best/1” strategy, which made full use of the optimal individual information in the neighborhood to guide the mutation operation. Zhang and Sanderson [19] proposed an adaptive differential evolution algorithm (JADE), which adopted “DE/current-to-pbest/1” mutation model, used suboptimal solutions to improve population diversity, and employed Cauchy and Normal distribution to generate F and CR. Qin et al. [20] proposed a self-adaptive DE (SaDE), which adopted four mutation strategies to generate mutation individuals. The selection of mutation strategy would be affected by previous performance. A DE algorithm (CoDE) using three mutation strategies and three parameters for the random combination was presented by Wang et al. [21]. Epitropakis et al. [22] proposed a novel framework that specified the selection probability in the mutation operation based on the distance between each individual and the mutation individual, thereby guiding the population to global optimization. Mallipeddi et al. [23] proposed the EPSDE algorithm, which was characterized by a stochastic selection of mutation strategies and parameters in a candidate pool consisting of three basic mutation strategies and preset parameters. Xiang et al. [24] proposed an enhanced differential evolution algorithm (EDE), which adopted a new combined mutation strategy composed of “DE/current/1” and “DE/pbest/1.” Cui et al. [25] proposed a DE algorithm based on adaptive multiple subgroups (MPADE), which divided the population into three subgroups according to fitness values, each subgroup had its mutation strategy. Wu et al. [26] presented a DE with multipopulation-based ensemble of mutation strategies (MPEDE), which had three mutation strategies, three indicator subgroups, and one reward subgroup. After several evolutionary generations, the reward subgroup was dynamically assigned to the best-performing mutation strategy. Parameters with an adaptive learning mechanism for the enhancement of differential evolution (PALM-DE) were presented by Meng et al. [27]. Unlike the external archive of the mutation strategy in JADE [19] and SHADE [9], the inferior solution archive in PALM-DE mutation strategy used a timestamp mechanism. In [28], Meng et al. introduced a novel parabolic population size reduction scheme and an enhanced timestamp-based mutation strategy to tackle the weakness of previous mutation strategy. Wei et al. [29] proposed the RPMDE algorithm, designed the “DE/M_pbest-best/1” mutation strategy, used the optimal individual group information to generate new solutions, and adopted the random perturbation method to avoid falling into the local optimal. Duan’s DPLDE [30] algorithm used population diversity and population fitness to determine individuals participating in mutation operation, thus influencing the mutation strategy. Tian and Gao [31] proposed NDE, which employed two mutation operators based on neighborhood-based and an individual-based selection probability to adjust the search performance of each individual appropriately. Wang et al. [32] proposed the DE algorithm based on particle swarm optimization (DEPSO), which utilized the improved “DE/rand/1” mutation strategy and PSO mutation strategy. Meng and Pan [33] presented hierarchical archive based on mutation strategy with depth information of evolution for the enhancement of differential evolution (HARD-DE), the depth information in which was the linkage of more than three different generations of populations and was included into the mutation strategy. A hybrid differential evolution algorithm based on “DE/target-to-ci_mbest/1” mutation operation of CIPDE [34] and “DE/target-to-pbest/1” mutation operation of JADE [19] was introduced by Pan et al. [35]. Meng et al. [36] proposed depth information-based DE with adaptive parameter control (Di-DE), the mutation strategy of which contained a depth information-based external archive.
As mentioned above, mutation strategies and control parameters affect the performance of DE, and “DE/rand/1” is widely used due to its strong global exploration ability and good population diversity. Many researchers have refined the mutation strategy. In this paper, an enhanced mutation strategy based on “DE/rand/1” is proposed by introducing a reference factor. Besides, according to the maximum, minimum, average fitness value of population, and the fitness value of the individual, the scaling factor and crossover probability are changed to adjust the population diversity effectively.
The remainder of the paper is organized as follows. Section 2 describes the basic DE algorithm. Section 3 provides the details of the proposed DSIDE. In Section 4, the proposed DSIDE is compared and analyzed experimentally with seven advanced DE algorithms, and the effectiveness of the enhanced mutation strategy and the novel adaptive strategy for control parameters in DSIDE is studied. Section 5 summarizes the work of this paper and points out the future research direction.
2. The Basic Differential Evolution Algorithm
2.1. Initialization
2.2. Mutation
2.3. Crossover
2.4. Selection
3. DSIDE Algorithm
In DSIDE, the crossover and selection operations are the same as the basic DE, as shown in equations (4) and (5), respectively. Next, the improved mutation strategy and adaptive strategy will be introduced.
3.1. An Enhanced Mutation Strategy
In equation (6), αi(∈[0,1]), Fi, and CRi are the reference factor, scaling factor, and crossover probability for each target individual , respectively; G denotes the current generation number. In equation (7), r means a random number on the interval [0,1]. Gmax represents the maximum generation number. From equation (7), it is not challenging to observe that the value of is relatively large at the initial evolutionary stage, which ensures a wide range of search. As the evolutionary generation increases, the value decreases and the search scope shrink.
3.2. A Novel Adaptive Strategy for Control Parameters
The reference factor , scaling factor , and crossover probability are updated before each evolution. The entire process of DSIDE algorithm is shown in Algorithm 1.
-
Algorithm 1: DSIDE.
- (1)
Initialize the original population pop and calculate their fitness values, NP = 100, G = 1, Gmax=1000;
- (2)
while ((G ≤ Gmax)do
- (3)
for each individual Xi in pop do
- (4)
Calculate αi in equation (7);
- (5)
Calculate Fi in equation (8);
- (6)
Calculate CRi in equation (9);
- (7)
Implement mutation in equation (6);
- (8)
Implement crossover in equation (4);
- (9)
Implement selection in equation (5);
- (10)
end for
- (11)
G = G + 1
- (12)
end while
4. Experimental Results and Analysis
4.1. Benchmark Functions
Unlike deterministic algorithms, it is difficult to verify that evolutionary algorithms are superior to other algorithms due to their limited knowledge. Therefore, benchmark functions are utilized to evaluate the performance of evolutionary algorithms. In this section, the performance of DSIDE is tested on 27 benchmark functions [37–39] listed in Table 1, where D is the dimension of the problem. f1 ~ f11 are unimodal functions. f12 has one minimum and is discontinuous. f13 is a noisy quadratic function. f14 ~ f27 are multimodal functions. f( ∗) denotes the global minimum value.
| Name | Function | Range | f( ∗) |
|---|---|---|---|
| Sphere | [−100,100]D | 0 | |
| Elliptic | [−100,100]D | 0 | |
| Bent cigar | [−100,100]D | 0 | |
| Schwefel 1.2 | [−100,100]D | 0 | |
| Schwefel 2.22 | [−10,10]D | 0 | |
| Schwefel 2.21 | f6(x) = max{|xi|, 1 ≤ i ≤ D} | [−100,100]D | 0 |
| Powell sum | [−100,100]D | 0 | |
| Sum squares | [−10,10]D | 0 | |
| Discuss | [−100,100]D | 0 | |
| Different powers | [−100,100]D | 0 | |
| Zakharov | [−5,10]D | 0 | |
| Step | [−100,100]D | 0 | |
| Noise quartic | [−1.28, 1.28]D | 0 | |
| Rosenbrock | [−30,30]D | 0 | |
| Griewank | [−600,600]D | 0 | |
| Rastrigin | [−5.12, 5.12]D | 0 | |
| Apline | [−100,100]D | 0 | |
| Bohachevsky_2 | [−100,100]D | 0 | |
| Salomon | [−100,100]D | 0 | |
| Scaffer2 | [−100,100]D | 0 | |
| Weierstrass | [−0.5, 0.5]D | 0 | |
| Katsuura | [−100,100]D | 0 | |
| HappyCat | [−100,100]D | 0 | |
| HGBat | [−100,100]D | 0 | |
| Scaffer’s F6 | [−0.5, 0.5]D | 0 | |
| Expanded Scaffer’s F6 | f26(x) = f25(x1, x2) + f25(x2, x3) + ⋯+f25(xD−1, xD) + f25(xD, x1) | [−5,5]D | 0 |
| Expanded Griewank’s plus Rosenbrock’s | f27(x) = f15(f14(x1, x2)) + f15(f14(x2, x3)) + ⋯+f15(f14(xD−1, xD)) + f15(f14(xD, x1)) | [−5.12, 5.12]D | 0 |
| NCRastrigin | [−10,10]D | 0 | |
| Levy and Montalvo 1 | [−10,10]D | 0 | |
| [−10,10]D | |||
| Levy and Montalvo 2 | [−5,5]D | 0 |
Experiment results in this paper are obtained on Windows 10 x64 Operating System of a PC with Intel (R) Core (TM) i7-8550U CPU (1.80 GHz) and 8 GB RAM, and algorithms are implemented in MATLAB 2015b Windows version.
4.2. Comparison with 7 Improved DE Algorithms
Here, we mainly discuss the overall optimization performance among jDE [6], JADE [19], SaDE [20], CoDE [21], EPSDE [23], MPEDE [26], DEPSO [32], and the proposed DSIDE algorithm. Experiments are carried out on f1 ~ f30 benchmark functions at 30 D and 100 D, respectively. The parameters of other algorithms are the same as in their original literatures. The population size NP is set to 100 for all algorithms. 30 independent runs with 1000 maximum number of evolutionary generations are conducted. Tables 2 and 3 show the mean/std (mean value and standard deviation) of fitness error over 30 runs at 30 D and 100 D, respectively. Symbols “+,” “≈,” and “−” behind “mean ± std” pair denote “Better Performance,” “Similar Performance,” and “Worse Performance,” respectively, all of which are measured under Wilcoxon’s signed-rank test with a level of significant α = 0.05. Furthermore, Wilcoxon’s rank-sum test and Kruskal–Wallis test [39, 40] in Tables 4–6 are employed to further test the optimization performance of all algorithms. The best results in tables are shown in bold. In addition, the representative convergence curves of all algorithms are also given in Figures 1 and 2.
| F | jDE | JADE | SaDE | CoDE | EPSDE | MPEDE | DEPSO | DSIDE |
|---|---|---|---|---|---|---|---|---|
| mean ± std | mean ± std | mean ± std | mean ± std | mean ± std | mean ± std | mean ± std | mean ± std | |
| f1 | 8.73E − 18 ± 7.55E − 18 | 4.55E − 36 ± 2.17E − 35 | 5.15E − 24 ± 4.88E − 24 | 5.96E − 08 ± 2.50E − 08 | 1.43E − 26 ± 4.42E − 26 | 1.46E − 29 ± 7.94E − 29 | 2.97E − 99 ± 1.50E − 98 | 0.00E + 00 ± 0.00E + 00 |
| f2 | 2.11E − 14 ± 1.51E − 14 | 2.79E − 31 ± 1.28E − 30 | 1.03E − 20 ± 1.53E − 20 | 4.19E − 05 ± 1.66E − 05 | 2.33E − 22 ± 3.90E − 22 | 3.77E − 27 ± 1.66E − 26 | 4.62E − 96 ± 2.52E − 95 | 0.00E + 00 ± 0.00E + 00 |
| f3 | 6.52E − 12 ± 4.85E − 12 | 1.03E − 30 ± 2.97E − 30 | 3.29E − 18 ± 2.78E − 18 | 3.93E − 02 ± 1.40E − 02 | 3.66E − 21 ± 9.09E − 21 | 1.70E − 25 ± 6.43E − 25 | 2.67E − 87 ± 1.46E − 86 | 0.00E + 00 ± 0.00E + 00 |
| f4 | 4.13E + 00 ± 5.42E + 00 | 1.26E − 13 ± 1.41E − 13 | 5.82E − 01 ± 4.56E − 01 | 6.99E − 02 ± 3.65E − 02 | 7.70E − 01 ± 4.18E + 00 | 1.35E − 06 ± 7.28E − 06 | 6.29E − 89 ± 3.25E − 88 | 0.00E + 00 ± 0.00E + 00 |
| f5 | 3.42E − 11 ± 1.34E − 11 | 1.10E − 14 ± 3.28E − 14 | 7.08E − 14 ± 3.16E − 14 | 5.00E − 05 ± 9.51E − 06 | 8.60E − 11 ± 1.21E − 10 | 7.62E − 16 ± 1.87E − 15 | 9.66E − 53 ± 3.75E − 52 | 0.00E + 00 ± 0.00E + 00 |
| f6 | 8.43E − 01 ± 8.20E − 01 | 8.93E − 12 ± 1.79E − 11 | 4.56E − 02 ± 7.63E − 02 | 3.20E − 01 ± 8.09E − 02 | 1.43E − 01 ± 1.44E − 01 | 1.56E − 07 ± 1.71E − 07 | 5.67E − 47 ± 3.11E − 46 | 0.00E + 00 ± 0.00E + 00 |
| f7 | 3.10E − 41 ± 1.55E − 40 | 9.36E − 44 ± 4.29E − 43 | 4.48E − 22 ± 1.81E − 21 | 2.14E − 17 ± 5.29E − 17 | 3.95E − 38 ± 2.05E − 37 | 6.15E − 53 ± 3.29E − 52 | 1.59E − 115 ± 8.70E − 115 | 0.00E + 00 ± 0.00E + 00 |
| f8 | 1.37E − 18 ± 1.36E − 18 | 2.92E − 37 ± 1.02E − 36 | 6.13E − 25 ± 5.88E − 25 | 6.68E − 09 ± 2.67E − 09 | 3.34E − 28 ± 5.36E − 28 | 4.89E − 32 ± 2.63E − 31 | 4.93E − 98 ± 2.70E − 97 | 0.00E + 00 ± 0.00E + 00 |
| f9 | 1.78E − 17 ± 2.03E − 17 | 1.43E − 32 ± 7.67E − 32 | 1.25E − 23 ± 1.07E − 23 | 8.20E − 08 ± 3.35E − 08 | 1.21E − 25 ± 2.64E − 25 | 9.10E − 29 ± 4.54E − 28 | 2.06E − 98 ± 1.01E − 97 | 0.00E + 00 ± 0.00E + 00 |
| f10 | 9.12E − 13 ± 5.83E − 13 | 1.07E − 23 ± 3.33E − 23 | 1.88E − 12 ± 2.77E − 12 | 5.14E − 06 ± 1.63E − 06 | 1.35E − 18 ± 1.90E − 18 | 1.13E − 20 ± 3.84E − 20 | 5.52E − 55 ± 2.89E − 54 | 0.00E + 00 ± 0.00E + 00 |
| f11 | 2.25E − 15 ± 3.04E − 15 | 1.27E − 39 ± 6.75E − 39 | 1.17E − 21 ± 1.14E − 21 | 2.06E − 08 ± 9.16E − 09 | 1.40E − 26 ± 5.53E − 26 | 7.92E − 34 ± 2.01E − 33 | 5.09E − 104 ± 1.33E − 103 | 0.00E + 00 ± 0.00E + 00 |
| f12 | 9.58E − 18 ± 1.04E − 17 | 3.08E − 34 ± 1.24E − 33 | 4.94E − 24 ± 6.24E − 24 | 6.13E − 08 ± 1.49E − 08 | 5.99E − 27 ± 1.74E − 26 | 2.12E − 31 ± 6.85E − 31 | 2.73E + 00 ± 2.69E − 01 | 1.18E + 00 ± 2.03E − 01 |
| f13 | 1.18E − 02 ± 3.27E − 03 | 3.98E − 03 ± 1.54E − 03 | 4.87E − 03 ± 1.87E − 03 | 1.38E − 02 ± 3.77E − 03 | 4.77E − 03 ± 2.22E − 03 | 3.44E − 03 ± 1.61E − 03 | 3.43E − 01 ± 2.39E − 01 | 8.49E − 04 ± 7.45E − 04 |
| f14 | 2.66E + 01 ± 1.36E + 01 | 1.65E − 01 ± 8.37E − 01 | 2.90E + 01 ± 1.44E + 01 | 2.23E + 01 ± 5.32E − 01 | 1.04E + 01 ± 3.21E + 00 | 3.13E + 00 ± 4.07E + 00 | 2.80E + 01 ± 2.77E − 01 | 2.85E + 01 ± 9.98E − 02 |
| f15 | 0.00E + 00 ± 0.00E + 00 | 1.78E − 11 ± 9.73E − 11 | 1.64E − 03 ± 4.26E − 03 | 1.82E − 05 ± 4.47E − 05 | 0.00E + 00 ± 0.00E + 00 | 1.40E − 03 ± 3.76E − 03 | 7.66E − 04 ± 4.20E − 03 | 0.00E + 00 ± 0.00E + 00 |
| f16 | 1.47E − 04 ± 7.04E − 04 | 1.87E − 04 ± 8.99E − 05 | 5.32E − 01 ± 8.10E − 01 | 2.46E + 01 ± 1.94E + 00 | 6.29E − 01 ± 7.87E − 01 | 2.78E − 12 ± 6.03E − 12 | 3.94E + 00 ± 2.14E + 01 | 0.00E + 00 ± 0.00E + 00 |
| f17 | 1.47E − 03 ± 4.72E − 04 | 1.13E − 02 ± 3.83E − 03 | 8.25E − 04 ± 3.77E − 04 | 3.09E + 01 ± 3.80E + 00 | 1.42E − 02 ± 5.22E − 03 | 2.27E − 07 ± 1.24E − 06 | 1.57E − 51 ± 4.78E − 51 | 0.00E + 00 ± 0.00E + 00 |
| f18 | 1.30E − 16 ± 2.11E − 16 | 0.00E + 00 ± 0.00E + 00 | 9.34E − 02 ± 3.08E − 01 | 4.32E − 06 ± 2.06E − 06 | 0.00E + 00 ± 0.00E + 00 | 3.98E − 02 ± 2.18E − 01 | 0.00E + 00 ± 0.00E + 00 | 0.00E + 00 ± 0.00E + 00 |
| f19 | 2.15E − 01 ± 3.45E − 02 | 2.03E − 01 ± 1.83E − 02 | 2.13E − 01 ± 3.46E − 02 | 3.64E − 01 ± 4.62E − 02 | 1.68E − 01 ± 4.68E − 02 | 2.57E − 01 ± 5.68E − 02 | 9.99E − 02 ± 1.53E − 07 | 0.00E + 00 ± 0.00E + 00 |
| f20 | 5.68E + 00 ± 1.32E + 00 | 4.63E + 00 ± 7.82E − 01 | 1.64E + 01 ± 2.68E + 00 | 2.05E + 01 ± 2.97E + 00 | 1.19E + 01 ± 1.68E + 00 | 2.26E + 00 ± 1.13E + 00 | 3.83E − 01 ± 1.01E + 00 | 0.00E + 00 ± 0.00E + 00 |
| f21 | 3.38E − 10 ± 3.98E − 10 | 2.40E − 05 ± 3.93E − 05 | 7.41E − 14 ± 8.66E − 14 | 1.26E − 02 ± 1.57E − 03 | 6.42E + 00 ± 2.36E + 00 | 4.38E − 03 ± 1.48E − 02 | 0.00E + 00 ± 0.00E + 00 | 0.00E + 00 ± 0.00E + 00 |
| f22 | 5.66E − 02 ± 8.32E − 03 | 2.65E − 03 ± 3.29E − 04 | 1.45E − 01 ± 1.92E − 02 | 8.84E − 03 ± 7.08E − 04 | 8.85E − 02 ± 1.32E − 02 | 6.99E − 04 ± 1.43E − 04 | 6.16E − 01 ± 9.64E − 02 | 0.00E + 00 ± 0.00E + 00 |
| f23 | 3.66E − 01 ± 5.20E − 02 | 2.53E − 01 ± 3.62E − 02 | 3.59E − 01 ± 6.11E − 02 | 4.85E − 01 ± 4.78E − 02 | 3.30E − 01 ± 3.86E − 02 | 2.76E − 01 ± 5.31E − 02 | 8.49E − 01 ± 9.02E − 02 | 5.04E − 01 ± 6.36E − 02 |
| f24 | 3.39E − 01 ± 3.77E − 02 | 3.71E − 01 ± 1.28E − 01 | 4.05E − 01 ± 1.15E − 01 | 2.96E − 01 ± 2.72E − 02 | 3.41E − 01 ± 7.43E − 02 | 4.11E − 01 ± 1.61E − 01 | 4.98E − 01 ± 6.20E − 03 | 4.88E − 01 ± 1.51E − 02 |
| f25 | 0.00E + 00 ± 0.00E + 00 | 0.00E + 00 ± 0.00E + 00 | 0.00E + 00 ± 0.00E + 00 | 2.43E − 12 ± 1.10E − 12 | 0.00E + 00 ± 0.00E + 00 | 0.00E + 00 ± 0.00E + 00 | 0.00E + 00 ± 0.00E + 00 | 0.00E + 00 ± 0.00E + 00 |
| f26 | 7.91E − 01 ± 1.37E − 01 | 6.77E − 01 ± 6.13E − 02 | 1.21E + 00 ± 1.46E − 01 | 1.70E + 00 ± 1.77E − 01 | 2.04E + 00 ± 2.45E − 01 | 3.45E − 01 ± 7.08E − 02 | 4.63E + 00 ± 8.45E − 01 | 0.00E + 00 ± 0.00E + 00 |
| f27 | 3.24E + 00 ± 3.45E − 01 | 2.83E + 00 ± 2.07E − 01 | 6.28E + 00 ± 5.62E − 01 | 8.47E + 00 ± 6.77E − 01 | 4.59E + 00 ± 4.34E − 01 | 2.11E + 00 ± 2.55E − 01 | 1.17E + 01 ± 4.46E − 01 | 1.18E + 01 ± 4.67E − 01 |
| f28 | 0.00E + 00 ± 0.00E + 00 | 0.00E + 00 ± 0.00E + 00 | 0.00E + 00 ± 0.00E + 00 | 8.00E + 00 ± 1.20E + 00 | 4.84E − 06 ± 2.39E − 05 | 0.00E + 00 ± 0.00E + 00 | 0.00E + 00 ± 0.00E + 00 | 0.00E + 00 ± 0.00E + 00 |
| f29 | 3.91E − 21 ± 3.66E − 21 | 1.57E − 32 ± 5.57E − 48 | 2.01E − 27 ± 2.69E − 27 | 6.64E − 11 ± 3.04E − 11 | 2.57E − 22 ± 9.18E − 22 | 1.57E − 32 ± 5.57E − 48 | 8.21E − 02 ± 3.15E − 02 | 2.78E − 03 ± 1.82E − 03 |
| f30 | 2.25E − 20 ± 3.06E − 20 | 5.83E − 29 ± 2.53E − 28 | 6.30E − 26 ± 7.49E − 26 | 1.52E − 10 ± 7.18E − 11 | 1.05E − 23 ± 2.34E − 23 | 9.40E − 30 ± 4.89E − 29 | 3.29E − 01 ± 7.20E − 02 | 6.19E − 01 ± 2.04E − 01 |
| − | 20 | 20 | 22 | 23 | 20 | 21 | 23 | |
| ≈ | 3 | 3 | 2 | 0 | 3 | 2 | 4 | |
| + | 7 | 7 | 6 | 7 | 7 | 7 | 3 |
| F | jDE | JADE | SaDE | CoDE | EPSDE | MPEDE | DEPSO | DSIDE |
|---|---|---|---|---|---|---|---|---|
| mean ± std | mean ± std | mean ± std | mean ± std | mean ± std | mean ± std | mean ± std | mean ± std | |
| f1 | 1.62E − 05 ± 7.35E − 06 | 3.01E − 16 ± 3.28E − 16 | 2.97E − 04 ± 1.36E − 04 | 7.40E − 01 ± 2.67E − 01 | 2.17E − 08 ± 2.20E − 08 | 9.24E − 10 ± 6.63E − 10 | 5.93E − 95 ± 2.86E − 94 | 0.00E + 00 ± 0.00E + 00 |
| f2 | 8.40E − 02 ± 5.79E − 02 | 4.17E − 11 ± 4.24E − 11 | 9.04E − 01 ± 5.30E − 01 | 2.14E + 03 ± 6.19E + 02 | 2.40E − 04 ± 2.59E − 04 | 1.46E − 04 ± 1.74E − 04 | 2.58E − 84 ± 1.41E − 83 | 0.00E + 00 ± 0.00E + 00 |
| f3 | 1.57E + 01 ± 7.77E + 00 | 1.21E − 09 ± 1.60E − 09 | 3.41E + 02 ± 2.25E + 02 | 7.25E + 05 ± 1.84E + 05 | 2.02E − 02 ± 1.25E − 02 | 5.18E − 03 ± 1.21E − 02 | 8.14E − 84 ± 4.46E − 83 | 0.00E + 00 ± 0.00E + 00 |
| f4 | 8.14E + 03 ± 2.80E + 03 | 2.13E + 02 ± 1.02E + 02 | 1.59E + 03 ± 3.08E + 02 | 1.42E + 03 ± 3.22E + 02 | 2.96E + 04 ± 3.07E + 04 | 2.89E + 02 ± 1.33E + 02 | 6.91E − 89 ± 3.76E − 88 | 0.00E + 00 ± 0.00E + 00 |
| f5 | 7.69E − 04 ± 3.31E − 04 | 3.26E − 08 ± 3.18E − 08 | 1.26E − 03 ± 1.75E − 03 | 1.17E + 00 ± 2.04E − 01 | 8.04E − 05 ± 1.17E − 04 | 3.84E − 03 ± 1.92E − 02 | 2.40E − 49 ± 1.07E − 48 | 0.00E + 00 ± 0.00E + 00 |
| f6 | 2.70E + 01 ± 4.36E + 00 | 7.18E + 00 ± 9.65E − 01 | 1.18E + 01 ± 1.82E + 00 | 5.00E + 00 ± 7.39E − 01 | 6.70E + 01 ± 1.87E + 01 | 1.00E + 01 ± 1.56E + 00 | 1.06E − 45 ± 3.44E − 45 | 0.00E + 00 ± 0.00E + 00 |
| f7 | 3.56E + 27 ± 1.80E + 28 | 5.29E + 33 ± 2.67E + 34 | 9.40E + 53 ± 5.13E + 54 | 8.33E + 20 ± 3.17E + 21 | 1.08E + 41 ± 5.36E + 41 | 1.41E + 38 ± 7.57E + 38 | 1.14E − 114 ± 6.23E − 114 | 0.00E + 00 ± 0.00E + 00 |
| f8 | 7.78E − 06 ± 3.71E − 06 | 2.06E − 16 ± 2.78E − 16 | 1.15E − 04 ± 5.85E − 05 | 2.89E − 01 ± 6.72E − 02 | 6.55E − 09 ± 3.47E − 09 | 8.29E − 10 ± 6.98E − 10 | 4.95E − 97 ± 2.64E − 96 | 0.00E + 00 ± 0.00E + 00 |
| f9 | 2.91E − 05 ± 1.29E − 05 | 9.47E − 16 ± 7.48E − 16 | 5.46E − 04 ± 2.94E − 04 | 9.39E − 01 ± 2.85E − 01 | 2.08E − 07 ± 1.08E − 07 | 6.02E − 09 ± 7.61E − 09 | 2.33E − 99 ± 6.98E − 99 | 0.00E + 00 ± 0.00E + 00 |
| f10 | 1.93E − 02 ± 3.38E − 02 | 2.56E − 08 ± 1.84E − 08 | 2.86E − 01 ± 9.05E − 02 | 1.49E + 00 ± 4.09E − 01 | 3.43E − 03 ± 2.80E − 03 | 1.23E − 05 ± 7.38E − 06 | 2.95E − 54 ± 1.16E − 53 | 0.00E + 00 ± 0.00E + 00 |
| f11 | 1.24E − 03 ± 7.08E − 04 | 8.30E − 16 ± 6.38E − 16 | 2.22E − 04 ± 1.27E − 04 | 1.86E − 02 ± 5.83E − 03 | 7.82E − 06 ± 8.22E − 06 | 2.63E − 09 ± 2.86E − 09 | 1.54E − 95 ± 8.42E − 95 | 0.00E + 00 ± 0.00E + 00 |
| f12 | 1.96E − 05 ± 9.68E − 06 | 2.58E − 16 ± 2.30E − 16 | 2.84E − 04 ± 1.37E − 04 | 7.84E − 01 ± 2.39E − 01 | 1.58E − 08 ± 8.52E − 09 | 9.56E − 10 ± 1.02E − 09 | 1.85E + 01 ± 5.49E − 01 | 1.19E + 01 ± 5.90E − 01 |
| f13 | 7.17E − 02 ± 1.28E − 02 | 4.21E − 02 ± 9.86E − 03 | 1.31E − 01 ± 2.88E − 02 | 6.42E − 02 ± 1.71E − 02 | 5.16E − 02 ± 2.20E − 02 | 7.10E − 02 ± 1.96E − 02 | 3.31E − 01 ± 1.86E − 01 | 9.70E − 04 ± 1.13E − 03 |
| f14 | 1.97E + 02 ± 5.74E + 01 | 1.47E + 02 ± 4.80E + 01 | 4.60E + 02 ± 8.04E + 01 | 2.07E + 02 ± 3.88E + 01 | 1.92E + 02 ± 5.14E + 01 | 1.77E + 02 ± 6.47E + 01 | 9.85E + 01 ± 2.12E − 01 | 9.83E + 01 ± 7.31E − 02 |
| f15 | 9.69E − 06 ± 4.04E − 06 | 2.46E − 03 ± 7.91E − 03 | 7.70E − 03 ± 1.24E − 02 | 4.16E − 01 ± 1.29E − 01 | 1.31E − 03 ± 3.59E − 03 | 4.92E − 03 ± 9.66E − 03 | 0.00E + 00 ± 0.00E + 00 | 0.00E + 00 ± 0.00E + 00 |
| f16 | 1.83E + 02 ± 1.53E + 01 | 1.56E + 02 ± 9.95E + 00 | 2.61E + 02 ± 1.33E + 01 | 7.14E + 02 ± 2.16E + 01 | 4.34E + 02 ± 2.70E + 01 | 4.35E + 01 ± 8.85E + 00 | 3.32E − 02 ± 1.82E − 01 | 0.00E + 00 ± 0.00E + 00 |
| f17 | 1.85E + 00 ± 2.21E + 00 | 1.58E + 01 ± 5.40E + 00 | 3.00E + 00 ± 4.15E + 00 | 8.86E + 01 ± 1.22E + 01 | 1.63E + 01 ± 2.63E + 01 | 2.49E + 00 ± 2.36E + 00 | 4.52E − 50 ± 1.48E − 49 | 0.00E + 00 ± 0.00E + 00 |
| f18 | 6.15E − 04 ± 3.10E − 04 | 2.35E + 00 ± 1.45E + 00 | 7.00E + 00 ± 2.71E + 00 | 2.06E + 01 ± 3.65E + 00 | 2.45E + 00 ± 2.02E + 00 | 5.08E + 00 ± 2.46E + 00 | 0.00E + 00 ± 0.00E + 00 | 0.00E + 00 ± 0.00E + 00 |
| f19 | 9.83E − 01 ± 8.78E − 02 | 7.00E − 01 ± 1.02E − 01 | 1.63E + 00 ± 2.27E − 01 | 1.90E + 00 ± 1.39E − 01 | 1.08E + 00 ± 1.84E − 01 | 1.39E + 00 ± 2.82E − 01 | 9.99E − 02 ± 1.54E − 07 | 0.00E + 00 ± 0.00E + 00 |
| f20 | 9.07E + 01 ± 9.50E + 00 | 8.89E + 01 ± 6.90E + 00 | 1.57E + 02 ± 2.02E + 01 | 3.01E + 02 ± 9.12E + 00 | 2.95E + 02 ± 2.55E + 01 | 1.25E + 01 ± 4.92E + 00 | 4.57E − 02 ± 1.44E − 01 | 0.00E + 00 ± 0.00E + 00 |
| f21 | 5.75E − 02 ± 1.55E − 02 | 2.79E + 00 ± 1.49E + 00 | 2.83E + 00 ± 1.32E + 00 | 6.06E + 00 ± 7.75E − 01 | 1.06E + 02 ± 2.95E + 00 | 1.35E + 01 ± 2.95E + 00 | 0.00E + 00 ± 0.00E + 00 | 0.00E + 00 ± 0.00E + 00 |
| f22 | 2.20E − 01 ± 2.56E − 02 | 1.08E − 01 ± 9.17E − 03 | 5.49E − 01 ± 3.87E − 02 | 4.01E − 01 ± 3.50E − 02 | 5.82E − 01 ± 3.74E − 02 | 9.35E − 03 ± 1.69E − 03 | 2.11E + 00 ± 1.80E − 01 | 0.00E + 00 ± 0.00E + 00 |
| f23 | 6.32E − 01 ± 5.38E − 02 | 5.17E − 01 ± 6.27E − 02 | 6.27E − 01 ± 7.99E − 02 | 7.20E − 01 ± 8.36E − 02 | 5.81E − 01 ± 5.89E − 02 | 5.76E − 01 ± 7.39E − 02 | 1.19E + 00 ± 1.38E − 01 | 8.72E − 01 ± 4.53E − 02 |
| f24 | 5.05E − 01 ± 1.73E − 01 | 5.03E − 01 ± 2.03E − 01 | 6.10E − 01 ± 1.98E − 01 | 6.39E − 01 ± 2.36E − 01 | 5.74E − 01 ± 1.96E − 01 | 5.46E − 01 ± 2.32E − 01 | 5.00E − 01 ± 6.71E − 11 | 4.99E − 01 ± 7.93E − 04 |
| f25 | 1.07E − 09 ± 7.45E − 10 | 1.48E − 16 ± 3.16E − 16 | 1.56E − 08 ± 8.66E − 09 | 3.55E − 05 ± 1.17E − 05 | 9.54E − 13 ± 6.18E − 13 | 1.02E − 13± 1.81E − 13 | 0.00E + 00 ± 0.00E + 00 | 0.00E + 00 ± 0.00E + 00 |
| f26 | 1.00E + 01 ± 8.76E − 01 | 9.01E + 00 ± 3.94E − 01 | 1.55E + 01 ± 6.55E − 01 | 2.28E + 01 ± 8.47E − 01 | 2.75E + 01 ± 1.97E + 00 | 2.36E + 00 ± 7.01E − 01 | 2.71E + 01 ± 1.09E + 01 | 0.00E + 00 ± 0.00E + 00 |
| f27 | 3.33E + 01 ± 3.12E + 00 | 2.88E + 01 ± 1.74E + 00 | 4.99E + 01 ± 2.06E + 00 | 7.00E + 01 ± 2.17E + 00 | 6.41E + 01 ± 7.46E + 00 | 1.84E + 01 ± 2.96E + 00 | 4.41E + 01 ± 3.76E − 01 | 4.43E + 01 ± 5.39E − 01 |
| f28 | 5.61E + 01 ± 1.07E + 01 | 5.03E + 01 ± 6.74E + 00 | 1.03E + 02 ± 1.17E + 01 | 6.24E + 02 ± 3.48E + 01 | 2.60E + 02 ± 2.70E + 01 | 3.32E + 01 ± 6.76E + 00 | 0.00E + 00 ± 0.00E + 00 | 0.00E + 00 ± 0.00E + 00 |
| f29 | 6.30E − 09 ± 4.06E − 09 | 1.95E − 20 ± 2.12E − 20 | 8.31E − 09 ± 3.84E − 09 | 6.26E − 05 ± 2.09E − 05 | 7.85E − 13 ± 8.09E − 13 | 2.07E − 03 ± 7.89E − 03 | 3.76E − 01 ± 6.85E − 02 | 2.23E − 01 ± 3.48E − 02 |
| f30 | 3.54E − 08 ± 2.69E − 08 | 3.16E − 17 ± 8.92E − 17 | 7.03E − 03 ± 1.15E − 02 | 2.24E − 03 ± 1.18E − 03 | 3.33E − 03 ± 5.17E − 03 | 9.99E − 03 ± 2.17E − 02 | 4.14E + 00 ± 6.81E − 01 | 7.38E + 00 ± 4.33E − 01 |
| − | 25 | 25 | 26 | 26 | 26 | 25 | 23 | |
| ≈ | 0 | 0 | 0 | 0 | 0 | 0 | 5 | |
| + | 5 | 5 | 4 | 4 | 4 | 5 | 2 |
| Comparison | R+ | D = 30 | p value | α = 0.1 | R+ | R− | p value | D = 100 | α = 0.05 | α = 0.1 |
|---|---|---|---|---|---|---|---|---|---|---|
| R− | α = 0.05 | |||||||||
| DSIDE vs. jDE | 235 | 143 | 6.37e − 04 | Yes | Yes | 372 | 93 | 6.01e − 06 | Yes | Yes |
| DSIDE vs. JADE | 223 | 155 | 1.53e − 03 | Yes | Yes | 371 | 94 | 6.94e − 06 | Yes | Yes |
| DSIDE vs. SaDE | 282 | 124 | 2.84e − 04 | Yes | Yes | 402 | 63 | 1.37e − 06 | Yes | Yes |
| DSIDE vs CoDE | 322 | 143 | 3.15e − 05 | Yes | Yes | 419 | 46 | 1.76e − 07 | Yes | Yes |
| DSIDE vs. EPSDE | 240 | 138 | 3.80e − 04 | Yes | Yes | 401 | 64 | 2.00e − 06 | Yes | Yes |
| DSIDE vs. MPEDE | 244 | 162 | 7.53e − 04 | Yes | Yes | 369 | 96 | 6.46e − 06 | Yes | Yes |
| DSIDE vs. DEPSO | 294 | 57 | 7.51e − 04 | Yes | Yes | 284 | 41 | 1.69e − 03 | Yes | Yes |
| Algorithms | jDE | JADE | SaDE | CoDE | EPSDE | MPEDE | DEPSO | DSIDE |
|---|---|---|---|---|---|---|---|---|
| Friedman (rank) | 5.25 | 3.12 | 5.42 | 6.73 | 4.92 | 3.53 | 4.28 | 2.75 |
| Kruskal–Wallis (rank) | 131.13 | 109.28 | 133.87 | 163.00 | 129.93 | 117.38 | 112.73 | 66.67 |
| Algorithms | jDE | JADE | SaDE | CoDE | EPSDE | MPEDE | DEPSO | DSIDE |
|---|---|---|---|---|---|---|---|---|
| Friedman (rank) | 4.80 | 3.10 | 6.23 | 6.77 | 5.50 | 4.23 | 3.32 | 2.05 |
| Kruskal–Wallis (rank) | 132.77 | 120.80 | 146.30 | 165.20 | 137.87 | 128.70 | 81.03 | 51.33 |
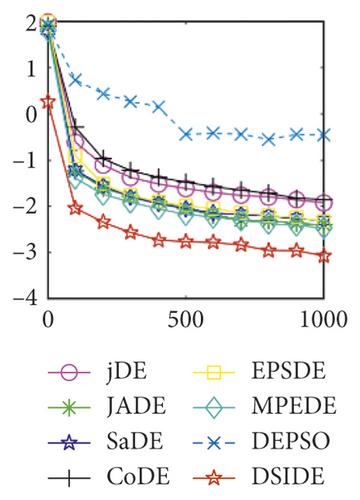
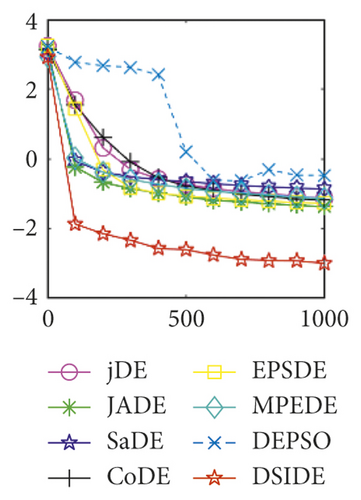
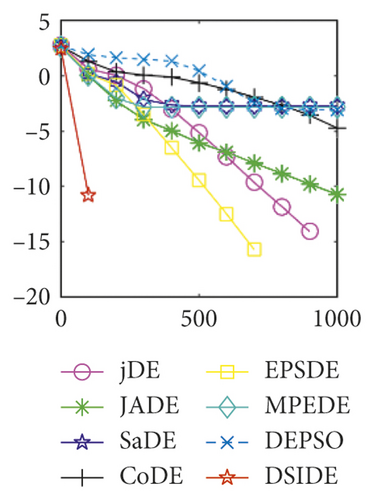
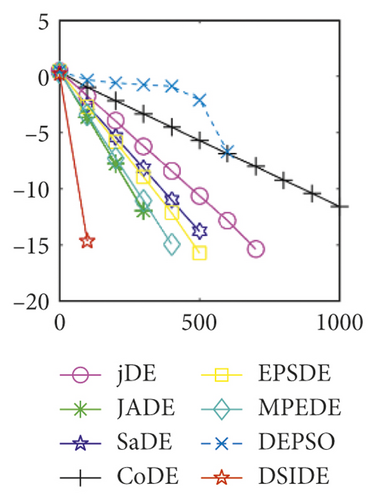
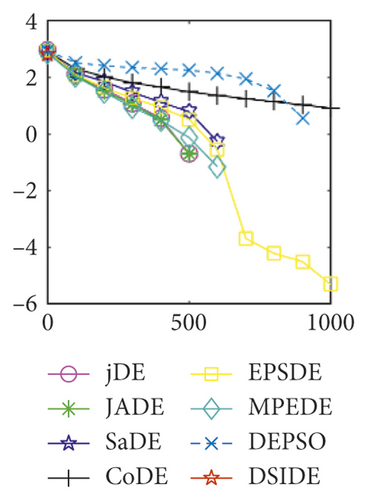
From Table 2 on 30 D, the proposed DSIDE algorithm displays that 23 out of 30 benchmark functions have better or similar performance than jDE, JADE, CoDE, EPSDE, and MPEDE, 24 out of 30 benchmark functions have better or comparable performance than SaDE, and 27 out of 30 benchmark functions have better or equivalent performance than DEPSO. Furthermore, the proposed DSIDE algorithm performs the best on benchmark functions f1 ~ f11, f13, f15 ~ f22, f25 ~ f26, and f28 in comparison with the other contrasted algorithms, performs slightly inferior on benchmark functions f14, f23, f24, and f27, and only performs poorly on f12, f29, and f30. Therefore, we can conclude that the proposed DSIDE algorithm is more competitive with the other seven improved DE algorithms on these functions at 30 D.
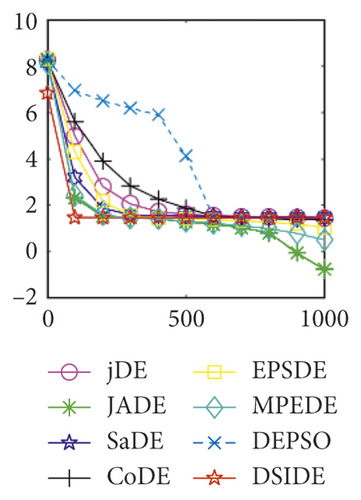
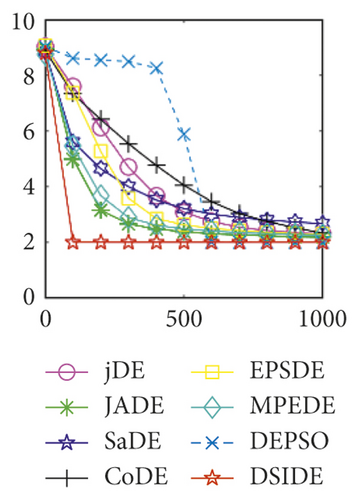
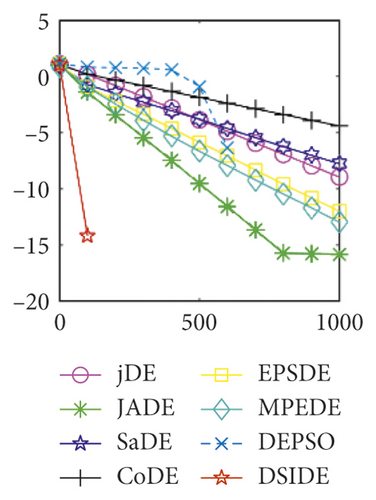
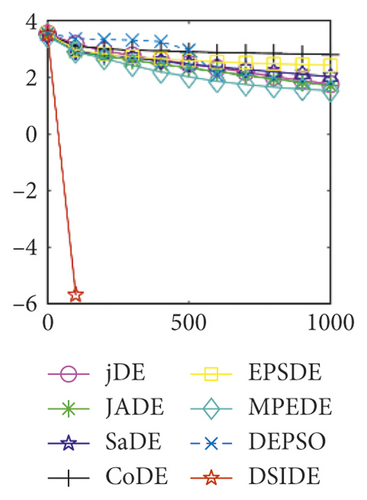
From Table 3 on 100 D, the proposed DSIDE algorithm displays that 25 out of 30 benchmark functions have better or equal performance than jDE, JADE, and MPEDE, 26 out of 27 benchmark functions have better or similar performance than SaDE, CoDE, and EPSDE, and 28 out of 30 benchmark functions have better or similar performance than DEPSO. Furthermore, the proposed DSIDE algorithm performs the best on benchmark functions f1 ~ f11, f13 ~ f22, f24 ~ f26, and f28 in comparison with all other contrasted algorithms, performs slightly inferior on benchmark functions f14, f23, f27, and only performs poorly on other three benchmark functions. That is to say, DSIDE has an overall better performance on benchmark functions f1 ~ f30 at 100 D.
From Table 4, we can see the results of Wilcoxon’s rank-sum test for 30 D and 100 D problems. R+ is the sum of positive ranks in which the first algorithm performs better than the second, and R− is the sum of negative ranks in which the first algorithm performs worse than the second. As shown in the table, we can observe that, for all comparison of DEs, all R+ values obtained by DSIDE are higher than R−. It proves that DSIDE outperforms other compared DE algorithms significantly. Tables 5 and 6, respectively, utilize Friedman and Kruskal–Wallis statistical test to compare the performance of each algorithm on 30 D and 100 D problems. It can be seen that the test results obtained by DSIDE are the minimum regardless of the high dimension or low dimension, indicating that DSIDE has the best performance among the comparison algorithms.
So far, all the nonparametric tests, including Wilcoxon’s rank-sum, Friedman, and Kruskal–Wallis test, support the conclusion that DSIDE is superior to other competing algorithms.
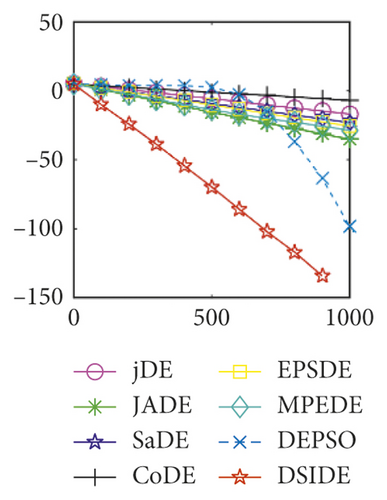
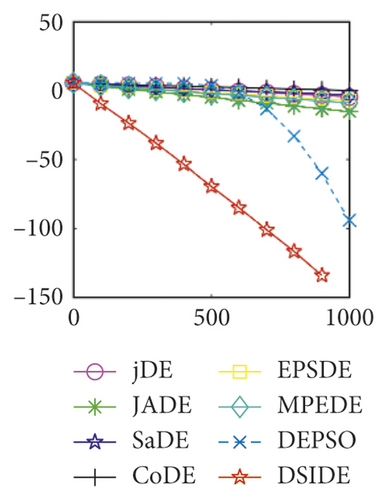
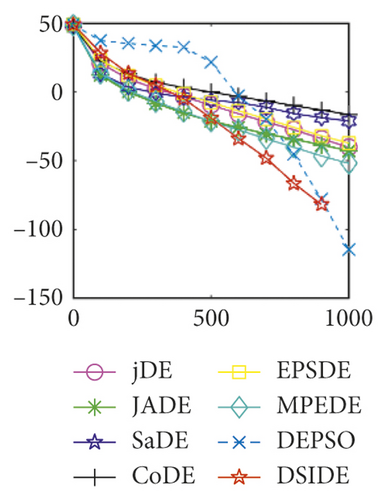
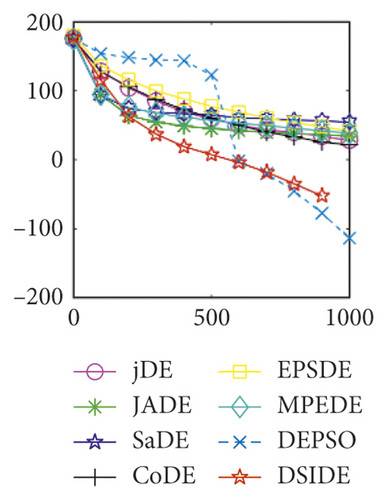
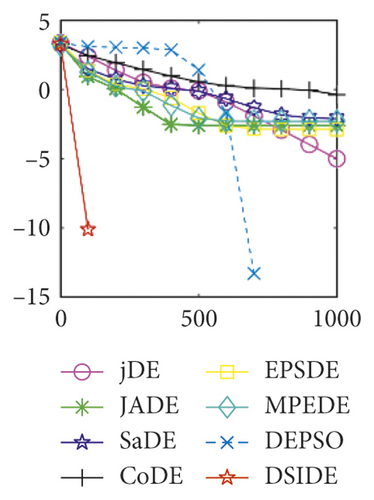
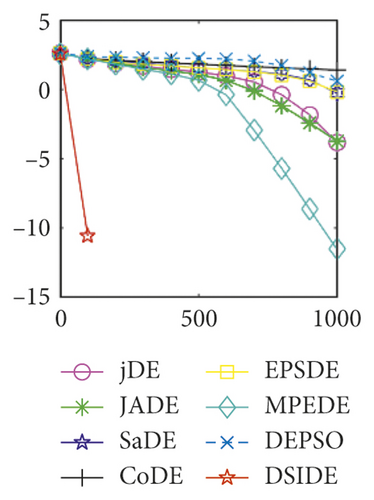
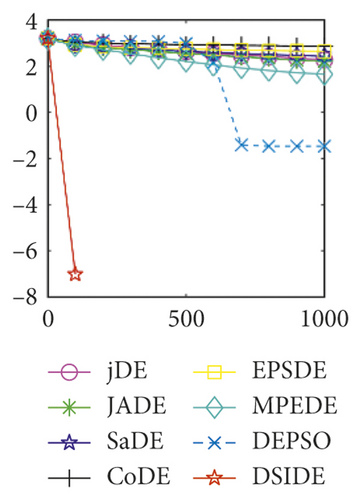
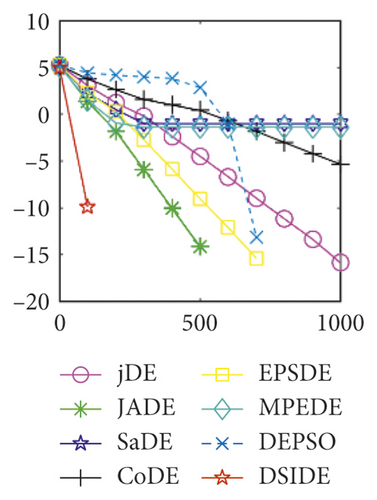
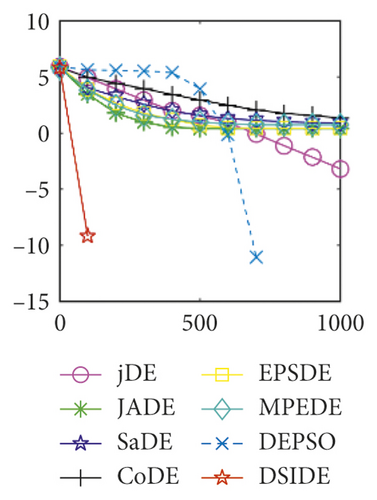
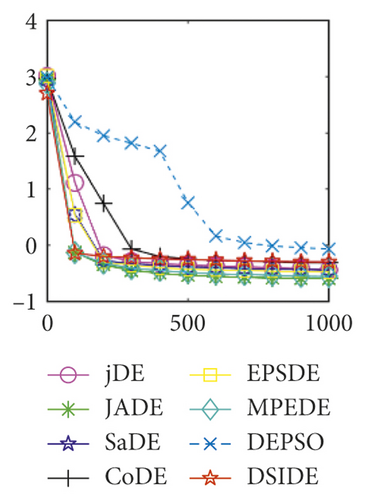
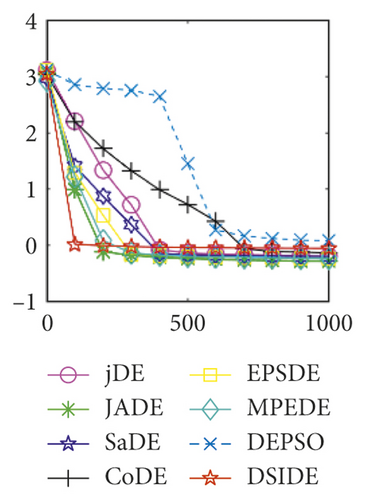
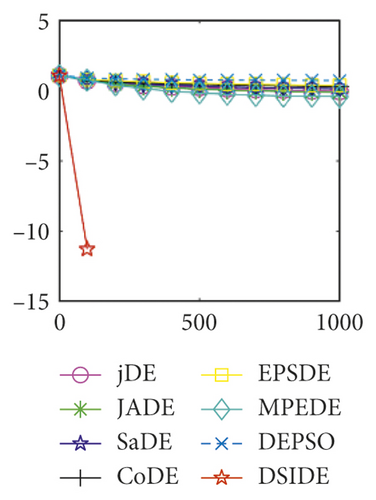
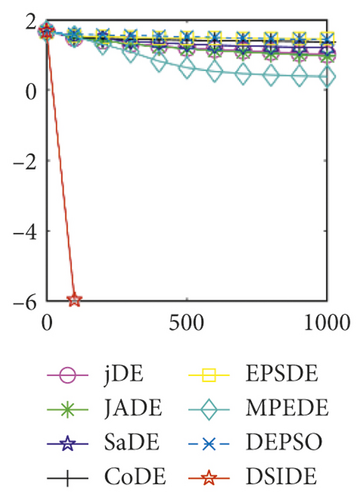
Furthermore, we compare the convergence curves of each algorithm on benchmark functions at 30 D and 100 D. All convergence curves are studied and analyzed from the aspects of convergence precision and whether they converge to the global optimum or not. Some representative convergence curves are depicted in Figures 1 and 2.
As shown in Figures 1(a) and 1(b), in convergence curves of function f1 at 30 D and 100 D, only DSIDE converges to the global optimum, and the average convergence accuracy is much higher than other algorithms under the same generations. Convergence curves of f7, as shown in Figures 1(c)and 1(d). Although convergence precision is not always optimal in the evolution process, only DSIDE gets the global optimum. Figures 1(e) and 1(f) show convergence curves of f13 at 30 D and 100 D, respectively. All algorithms have not found the optimal solution, but the average convergence accuracy of DSIDE is much higher than other algorithms under the same generations and obtains the best value. Figures 1(g) and 1(h) show convergence curves of f14 at 30 D and 100 D, respectively. All algorithms have not obtained the global minimum. JADE performs the best on the low-dimensional problem, while DSIDE is the best on high-dimensional. In Figures 1(i) and 1(j), DSIDE converges the fastest on f15. DSIDE, EPSDE, and jDE converge to the global optimum at 30 D, while DSIDE and DEPSO reach the optimal at 100 D. In Figures 1(k) and 1(l), only DSIDE gets the global optimal on f16 and consumes fewer generations and converges quickly.
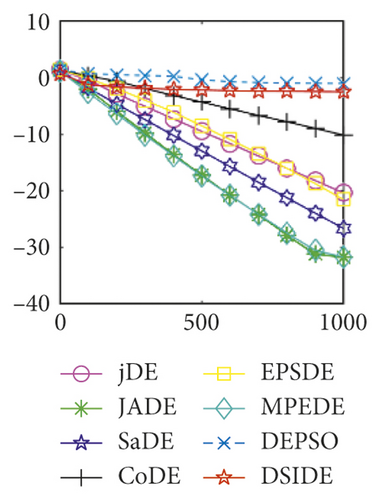
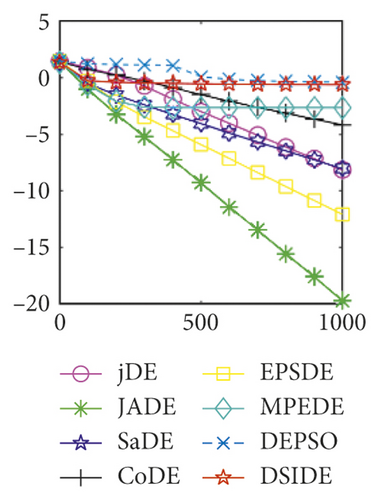
In Figure 2(a), DSIDE, JADE, DEPSO, and EPSDE obtain the optimal on f18 at 30 D. In Figure 2(b), DSIDE and DEPSO get the global optimal on f18 at 100D. DSIDE has the fastest convergence speed in both low-dimensional and high-dimensional problems. Convergence curves of function f23 in Figures 2(c) and 2(d), none of the algorithms finds the global minimum, and there is a phenomenon of “evolutionary stagnation.” In Figures 2(e) and 2(f) on function f25, only CoDE cannot find the global minimum at 30D; DSIDE and DEPSO get the global optimal at 100D, but the former costs much less generations. In Figures 2(g) and 2(h), DSIDE converges to the global optimal on f26, while other algorithms suffer from “evolutionary stagnation.” In Figure 2(i), the global minimum value is found by all algorithms except CoDE and EPSDE on f28 at 30 D. In Figure 2(j), DSIDE and DEPSO get the global optimal on function f28 at 100 D. In Figures 2(k) and 2(l), DSIDE performs relatively low but consistently outperforms DEPSO on function f29.
In general, through the comparative analysis of the above experiments, DSIDE not only obtains the global optimal value most times on these benchmark functions but also is superior to other algorithms in terms of convergence speed and convergence accuracy.
4.3. Efficiency Analysis of Proposed Algorithmic Components
- (i
)To verify the effectiveness of the enhanced mutation strategy of reference factor α, DSIDE variants adopt dynamic F, CR, and constant reference factor of α = 0.3 and α = 0.6 and random real number in [0,1], which are, respectively, called as DSIDE-1, DSIDE-2, and DSIDE-3 one by one.
- (ii)
To investigate the validity of the scaling factor adaptive strategy, DSIDE variants employ dynamic CR, α and fixed scaling factor of F = 0.3, F = 0.6, and random real number in [0,1], which are named DSIDE -4, DSIDE -5, and DSIDE -6 for short.
- (iii)
To study the contribution of the crossover probability adaptive strategy, DSIDE variants with shifty F, α and settled crossover probability of CR = 0.3 and CR = 0.6, and random real number in [0,1] are, respectively, abbreviated as DSIDE-7, DSIDE-8, and DSIDE-9.
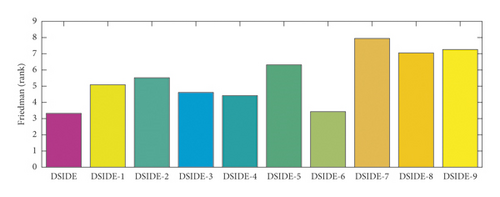
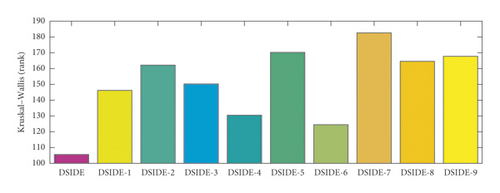
For the purpose of evaluating and comparing the performance of DSIDE variants, Friedman test, Kruskal–Wallis test, and Wilcoxon’s rank-sum test are adopted, and the test results are shown in Figure 3(a), Figure 3(b), and Table 7, respectively. The following summaries can be obtained. (1) From Figure 3, we can observe that DSIDE and DSIDE-6 are the best and the second, while the performance of other DSIDE variants is relatively low. The combined effect of the proposed algorithmic components is the best. (2) From Table 7, the integrated DSIDE performs significantly better than DSIDE variants (DSIDE-2 and DSIDE-5) with a larger reference factor and a lager scaling factor, as well as DSIDE variants (DSIDE-7, DSIDE-8, and DSIDE-9) with different crossover probability. The performance between the integrated DSIDE and DSIDE-1 with a smaller reference factor, DSIDE-3 with a random reference factor, and DSIDE-4 with a smaller scaling factor show no significant difference when the significance level of Wilcoxon’s rank-sum test is 0.1, but the difference is opposite when the significant level is 0.05. At the same time, there is no performance difference between DSIDE and DSIDE-6 with a random scaling factor, regardless of the significance level. The validity of the proposed mutation strategy and adaptive strategy for control parameters is demonstrated utilizing above experimental comparisons. It is noted that the contribution of the adaptive strategy of crossover probability is larger than enhanced mutation strategy and adaptive strategy of scaling factor. That is to say, although the enhanced mutation strategy of reference factor and adaptive strategy of scaling factor are effective, DSIDE is less susceptible to both a smaller or variational reference factor and scaling factor.
| Comparison | D = 30 | ||||
|---|---|---|---|---|---|
| R+ | R− | p value | α = 0.05 | α = 0.1 | |
| DSIDE vs. DSIDE-1 | 216 | 37 | 1.12e − 02 | No | Yes |
| DSIDE vs. DSIDE-2 | 165 | 135 | 5.63e − 03 | Yes | Yes |
| DSIDE vs. DSIDE-3 | 154 | 122 | 1.34e − 02 | No | Yes |
| DSIDE vs. DSIDE-4 | 75 | 30 | 2.21e − 01 | No | No |
| DSIDE vs. DSIDE-5 | 194 | 106 | 5.63e − 03 | Yes | Yes |
| DSIDE vs. DSIDE-6 | 59 | 46 | 3.00e − 01 | No | No |
| DSIDE vs. DSIDE-7 | 242 | 34 | 2.94e − 03 | Yes | Yes |
| DSIDE vs DSIDE-8 | 276 | 0 | 6.46e − 03 | Yes | Yes |
| DSIDE vs DSIDE-9 | 276 | 0 | 7.45e − 03 | Yes | Yes |
5. Conclusions
DSIDE’s innovation lies in two strategies, the enhanced mutation strategy and the novel adaptive strategy for control parameters. On the one hand, the enhanced mutation strategy considers the influence of the reference individual on the overall evolution. It introduces the reference factor, which is beneficial to global exploration in the early stage of evolution and global convergence in the later stage. On the other hand, the novel adaptive strategy for control parameters can dynamically adjust the scaling factor and crossover probability according to the fitness value, which has a positive impact on maintaining the population diversity. DSIDE is compared with other seven DE algorithms, the results are evaluated by three nonparametric statistical tests, and the convergence curves are analyzed. Experimental results show that the proposed DSIDE can effectively improve the optimization performance. Besides, the efficiency analysis of proposed algorithmic components has been carried out, which further proves the comprehensive effect and validity of DSIDE.
So far, DE variants have been applied to various fields, such as target allocation [41], text classification [42], image segmentation [43], and neural network [44–47]. For the future work, the proposed DSIDE algorithm will be applied to the parameter optimization of neural network and may further apply it to the air traffic control system for flight trajectory prediction [48, 49].
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China, no. U1833128.
Open Research
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.




