Toxoplasmosis and Polygenic Disease Susceptibility Genes: Extensive Toxoplasma gondii Host/Pathogen Interactome Enrichment in Nine Psychiatric or Neurological Disorders
Abstract
Toxoplasma gondii is not only implicated in schizophrenia and related disorders, but also in Alzheimer′s or Parkinson′s disease, cancer, cardiac myopathies, and autoimmune disorders. During its life cycle, the pathogen interacts with ~3000 host genes or proteins. Susceptibility genes for multiple sclerosis, Alzheimer′s disease, schizophrenia, bipolar disorder, depression, childhood obesity, Parkinson′s disease, attention deficit hyperactivity disorder (P from 8.01E − 05 (ADHD) to 1.22E − 71) (multiple sclerosis), and autism (P = 0.013), but not anorexia or chronic fatigue are highly enriched in the human arm of this interactome and 18 (ADHD) to 33% (MS) of the susceptibility genes relate to it. The signalling pathways involved in the susceptibility gene/interactome overlaps are relatively specific and relevant to each disease suggesting a means whereby susceptibility genes could orient the attentions of a single pathogen towards disruption of the specific pathways that together contribute (positively or negatively) to the endophenotypes of different diseases. Conditional protein knockdown, orchestrated by T. gondii proteins or antibodies binding to those of the host (pathogen derived autoimmunity) and metabolite exchange, may contribute to this disruption. Susceptibility genes may thus be related to the causes and influencers of disease, rather than (and as well as) to the disease itself.
1. Introduction
The protozoan parasite Toxoplasma gondii (T. gondii) which causes toxoplasmosis, is primarily hosted not only in cats but also in mice, rabbits, dogs, farmyard and wild animals, and domestic fowl, and is transmissible to man [1–5]. It has been implicated in the pathogenesis of many diseases, most notably schizophrenia [6–8], but also with bipolar disorder [9] depression and suicide attempts [10]. There is also evidence from serological antibody studies that the parasite may be implicated in the aetiology of Alzheimer’s and Parkinson’s disease [11–13] and in certain epilepsies of unknown origin [14]. The parasite has also been implicated in a number of autoimmune disorders including antiphospholipid syndrome, cryoglobulinemia, ANCA-associated vasculitides, autoimmune thyroid diseases, systemic sclerosis, rheumatoid arthritis, inflammatory bowel disease, and systemic lupus erythematosus, possibly related to host/pathogen antigen homology [15, 16].
It has already been noted that several schizophrenia susceptibility genes are related to the T. gondii life cycle, as well as to that of other pathogens implicated in this condition (cytomegalovirus, influenza, rubella, and herpes viruses) [17, 18] and that in both Alzheimer’s disease (herpes simplex, Chlamydia pneumoniae, Helicobacter pylori, and Cryptococcus neoformans) [19, 20] and multiple sclerosis (Epstein-Barr virus) [21], susceptibility genes are also related to the life cycles of suspect pathogens. In animal models, and without the aid of any gene variant, such agents can, per se, induce pathological features relevant to the disease process, for example amyloid deposition and tau phosphorylation (induced by herpes simplex, C. Pneumoniae, treponemas, Borrelia burgdorferi, and other spirochetes) [22–24], demyelination induced by various viruses [25], or dopaminergic overactivity in the case of T. gondii [26]. The H1N1strain of the influenza virus is also able to destroy neurones in the substantia nigra, provoking Parkinsonian symptoms in laboratory models [27]. Pathogens can thus be regarded as potential causes, likely acting in a gene dependent manner. Many such agents show a seroprevalence far above the incidence of the disease with which they are implicated; for example, T. gondii may infect 30% of the world’s population [28] in comparison to a schizophrenia prevalence of ~1% [29], and, as is the case with genetic risk factors, conflicting epidemiological data have often cast doubt upon whether such pathogens can truly cause disease [30]. However, this situation also applies to Helicobacter pylori, which indubitably causes stomach ulcers and likely gastric cancer [31, 32], although not all of the many infected with this agent (~50% of the world population [33]) succumb to these conditions. Any causative effects of such agents in man must therefore be conditioned by other factors, among which are immunity and resistance to the pathogen; pathogen strain or the timing and severity of infection; other confounding environmental and medical factors as well as the susceptibility genes for each disease. The effects of risk promoting gene variants, which are also present in control populations, albeit in lower proportion, must also be conditioned by environmental and epigenetic factors, as well as by gene/gene interactions.
During its life cycle any pathogen interacts with hundreds of human proteins whose function can only be compromised by their diversion to the attentions of the invader. In addition, bacteria and parasites scavenge important metabolites from host cells or fluids and donate other compounds to the host which must react accordingly. Activation of the immune system and inflammatory defence, involving chemokines, cytokines and numerous other mediators are an evident consequence of any infection, as are the resulting fevers [34]. It has also been noted in many bioinformatics studies that pathogen proteins closely resemble our own, and that immune attack directed towards the pathogen may thus result in antibody cross-reactivity with human proteins. The development of pathogen-derived autoantibodies may also play a key role in this pathological scenario [18, 19, 21, 35–39].
As shown below, the hundreds of human proteins implicated in the T. gondii life cycle are highly enriched in the products of susceptibility genes for the numerous conditions with which this parasite has been associated, as well as for others where a link is not yet suspected. The human pathways deranged by the parasite are also relevant to each condition. Subsets of the extensive T. gondii host/pathogen interactome appear to be relatively specific for distinct diseases suggesting that they relate to the cause of the disease, and that they may be able to direct the attentions of the pathogen towards particular pathways, pathologies, and disease.
2. Methods
Briefly, lists of several hundred susceptibility genes involved in eleven different diseases were compared with a list of several thousand host genes implicated in the T. gondii host/pathogen interactome. Any significant enrichment of interactome genes within susceptibility gene datasets (and vice versa) was identified by statistical analysis.
The genes and environmental factors implicated in the various diseases (Alzheimer’s disease, attention deficit hyperactivity disorder, autism, bipolar disorder, chronic fatigue syndrome, depression, schizophrenia, multiple sclerosis, Parkinson’s disease, anorexia, and childhood obesity) are listed at PolygenicPathways (http://www.polygenicpathways.co.uk/) and at sites therein (including the autism database at Mindspec (AutDB) [40], the Bipolar database at the University of Chicago [41], AlzGene, MSGene, PDGene and SZGene [42–45]). Genome-wide association data can be accessed at the National Human Genome Research Institute http://www.genome.gov/gwastudies/ [46].
Host/pathogen interactions for T. gondii and microarray data (mRNA expression changes in response to T. gondii infection) were collected by literature survey and are listed at http://www.polygenicpathways.co.uk/tgondii.htm. Pathway analysis of the human arm of this interactome was performed using KEGG mapper [47] http://www.genome.jp/kegg/tool/map_pathway2.html, and the results are posted at http://www.polygenicpathways.co.uk/keggtgondii.htm. These and various other files relating to the analysis are posted at http://www.polygenicpathways.co.uk/toxoplasmosis.htm.
3. Statistics
The human genome currently contains 26,846 genes, 2792 of which are contained in the T. gondii host/pathogen interactome. In any other dataset, one would expect 2792/26846 genes to be involved with the pathogen (10.4%). Similarly, for N susceptibility genes in any disorder, one would expect N/26846 to appear in the host/pathogen interactome, providing the expected numbers in each colliding dataset. The significance of differences between the observed and expected values was assessed using the chi-squared test. Statistical analysis for the enrichment of particular KEGG pathways within datasets was performed using the tools at the Consensus Path Database (CPDB) [48] developed by the Max Planck Institute for molecular genetics http://cpdb.molgen.mpg.de/CPDB. Overlapping gene sets were identified using the Venny tool [49] at http://bioinfogp.cnb.csic.es/tools/venny/index.
4. Results
4.1. KEGG Pathway Analysis of the T. gondii/Host Interactome
2792 proteins or mRNAs are involved in the host/pathogen interactome, approximating to 10% of the human genome. A summary of the KEGG pathway analysis of the human arm of this interactome is provided in Tables 1 and 2.
Number of genes |
P value | |
|---|---|---|
| Immune and defence | ||
| Cytokine-cytokine receptor interaction | (103) | 2.02E − 20 |
| Chemokine signalling pathway | (64) | 3.19E − 09 |
| Toll-like receptor signalling pathway | (52) | 9.24E − 17 |
| Phagosome | (47) | 1.02E − 05 |
| Natural killer cell mediated cytotoxicity | (46) | 2.73E − 07 |
| T cell receptor signalling pathway | (45) | 1.52E − 10 |
| Hematopoietic cell lineage | (42) | 1.52E − 12 |
| Leukocyte transendothelial migration | (36) | 0.000149 |
| NOD-like receptor signalling pathway | (34) | 9.21E − 14 |
| Fc epsilon RI signalling pathway | (29) | 0.000764 |
| Fc gamma R-mediated phagocytosis | (29) | 0.00115 |
| Complement and coagulation cascades | (28) | 5.80E − 07 |
| B cell receptor signalling pathway | (28) | 1.01E − 05 |
| Lysosome | (27) | |
| Antigen processing and presentation | (26) | 5.34E − 05 |
| Salivary secretion | (26) | |
| Adipocytokine signalling pathway | (25) | 0.000221 |
| RIG-I-like receptor signalling pathway | (23) | 0.000354 |
| Cytosolic DNA-sensing pathway | (22) | 0.000106 |
| Intestinal immune network for IgA production | (22) | 7.87E − 07 |
| Diseases | ||
| Pathways in cancer | (94) | 2.67E − 08 |
| Transcriptional misregulation in cancer | (52) | 3.32E − 06 |
| Prostate cancer | (34) | 6.94E − 06 |
| Small cell lung cancer | (31) | 4.04E − 06 |
| Colorectal cancer | (23) | 3.33E − 05 |
| Pancreatic cancer | (21) | 0.0019 |
| Acute myeloid leukaemia | (21) | 8.14E − 05 |
| Chronic myeloid leukaemia | (20) | 0.00747 |
| Glioma | (19) | 0.00964 |
| Renal cell carcinoma | (17) | |
| Endometrial cancer | (16) | 0.0048 |
| Non-small cell lung cancer | (15) | |
| Melanoma | (15) | |
| Bladder cancer | (14) | 0.00364 |
| Neurological | ||
| Alzheimer′s disease | (52) | 2.02E − 05 |
| Huntington′s disease | (34) | |
| Amyotrophic lateral sclerosis (ALS) | (30) | 7.94E − 10 |
| Parkinson′s disease | (27) | |
| Prion diseases | (22) | 2.26E − 08 |
| Autoimmune and atopicdiseases | ||
| Systemic lupus erythematosus | (45) | 2.48E − 06 |
| Rheumatoid arthritis | (44) | 1.26E − 12 |
| Type I diabetes mellitus | (25) | 1.11E − 08 |
| Allograft rejection | (23) | 1.56E − 09 |
| Autoimmune thyroid disease | (22) | 1.62E − 05 |
| Graft-versus-host disease | (21) | 3.63E − 06 |
| Asthma | (14) | 0.00422 |
| Primary immunodeficiency | (13) | 0.00168 |
| Cardiac | ||
| Viral myocarditis | (31) | 1.24E − 08 |
| Dilated cardiomyopathy | (27) | 0.00109 |
| Hypertrophic cardiomyopathy (HCM) | (25) | 0.00164 |
| Arrhythmogenic right ventricular cardiomyopathy (ARVC) | (18) | |
| Other | ||
| Alcoholism | (40) | |
| Type II diabetes mellitus | (19) | 0.00194 |
| Maturity onset diabetes of the young | (2) | |
| Other infections | ||
| HTLV-I infection | (85) | 1.62E − 10 |
| Tuberculosis | (80) | 6.67E − 19 |
| Influenza A | (69) | 5.11E − 14 |
| Toxoplasmosis | (66) | 6.90E − 20 |
| Herpes simplex infection | (66) | 6.88E − 12 |
| Epstein-Barr virus infection | (65) | |
| Measles | (56) | 3.36E − 13 |
| Amoebiasis | (56) | 4.59E − 13 |
| Chagas disease (American trypanosomiasis) | (55) | 2.70E − 16 |
| Pertussis | (52) | 1.38E − 20 |
| Leishmaniasis | (51) | 2.49E − 20 |
| Salmonella infection | (47) | 1.85E − 14 |
| Hepatitis C | (39) | 0.000135 |
| Legionellosis | (35) | 9.83E − 15 |
| Malaria | (32) | 2.90E − 13 |
| Shigellosis | (29) | 4.94E − 09 |
| Epithelial cell signalling in | ||
| Helicobacter pylori infection | (26) | 1.83E − 05 |
| Bacterial invasion of epithelial cells | (26) | 1.01E − 05 |
| African trypanosomiasis | (24) | 1.56E − 07 |
| Staphylococcus aureus infection | (24) | 7.63E − 07 |
| Pathogenic Escherichia coli infection | (21) | 0.000147 |
| Vibrio cholerae infection | (18) | |
| Number of genes | P value | |
|---|---|---|
| Signalling networks | ||
| MAPK signalling pathway | (72) | 8.43E − 06 |
| Jak-STAT signalling pathway | (59) | 9.01E − 12 |
| Calcium signalling pathway | (44) | 0.00797 |
| Insulin signalling pathway | (34) | |
| Wnt signalling pathway | (31) | |
| PPAR signalling pathway | (29) | 1.36E − 05 |
| GnRH signalling pathway | (28) | |
| ErbB signalling pathway | (26) | 0.00146 |
| p53 signalling pathway | (25) | 1.83E − 05 |
| VEGF signalling pathway | (24) | 0.0017 |
| TGF-beta signalling pathway | (19) | |
| Phosphatidylinositol signalling system | (19) | |
| mTOR signalling pathway | (15) | |
| Hedgehog signalling pathway | (7) | |
| Notch signalling pathway | (6) | |
| Tissue process | ||
| Osteoclast differentiation | (61) | 7.00E − 17 |
| Vascular smooth muscle contraction | (27) | |
| Bile secretion | (25) | |
| Melanogenesis | (25) | |
| Pancreatic secretion | (23) | |
| Mineral absorption | (22) | |
| Oocyte meiosis | (21) | |
| Carbohydrate digestion and absorption | (20) | 0.00207 |
| Protein digestion and absorption | (20) | |
| Endocrine and other factor-regulated calcium reabsorption | (19) | |
| Olfactory transduction | (17) | |
| Gastric acid secretion | (17) | |
| Aldosterone-regulated sodium reabsorption | (16) | 0.00283 |
| Progesterone-mediated oocyte maturation | (16) | |
| Cardiac muscle contraction | (13) | |
| Proximal tubule bicarbonate reclamation | (10) | |
| Vasopressin-regulated water reabsorption | (9) | |
| Taste transduction | (6) | |
| Vitamin digestion and absorption | (6) | |
| Collecting duct acid secretion | (4) | |
| Fat digestion and absorption | (4) | |
| Dorso-ventral axis formation | (4) | |
| Primary bile acid biosynthesis | (2) | |
| Renin-angiotensin system | (1) | |
| Cellular process | ||
| Focal adhesion | (56) | 5.95E − 06 |
| Cell adhesion molecules (CAMs) | (50) | 5.84E − 10 |
| Regulation of actin cytoskeleton | (49) | 0.00475 |
| Apoptosis | (45) | 1.75E − 15 |
| Endocytosis | (42) | |
| Protein processing in endoplasmic reticulum | (33) | |
| Extracellular matrix-receptor interaction | (31) | 2.29E − 06 |
| ABC transporters | (23) | |
| Gap junction | (22) | |
| Cell cycle | (20) | |
| Ubiquitin mediated proteolysis | (20) | |
| Tight junction | (18) | |
| RNA transport | (16) | |
| Adherens junction | (16) | |
| Peroxisome | (15) | |
| Ribosome | (14) | |
| Regulation of autophagy | (11) | |
| Ribosome biogenesis in eukaryotes | (10) | |
| Spliceosome | (10) | |
| Proteasome | (10) | |
| RNA degradation | (8) | |
| RNA polymerase | (8) | |
| Base excision repair | (8) | |
| Nucleotide excision repair | (6) | |
| DNA replication | (5) | |
| Circadian rhythm: mammal | (4) | |
| Basal transcription factors | (3) | |
| Protein export | (3) | |
| mRNA surveillance pathway | (3) | |
| Mismatch repair | (2) | |
| SNARE interactions in vesicular transport | (2) | |
| Homologous recombination | (1) | |
| Metabolism | ||
| Purine metabolism | (53) | 0.000397 |
| Pyrimidine metabolism | (31) | 0.00305 |
| Arginine and proline metabolism | (24) | 0.005 |
| Glycolysis/Gluconeogenesis | (23) | 0.0002 |
| Glutathione metabolism | (21) | 0.0008 |
| Arachidonic acid metabolism | (20) | |
| Glycerophospholipid metabolism | (20) | |
| Tryptophan metabolism | (19) | 0.002 |
| Oxidative phosphorylation | (19) | |
| Amino sugar and nucleotide sugar metabolism | (18) | |
| Inositol phosphate metabolism | (14) | |
| Fatty acid metabolism | (14) | |
| Galactose metabolism | (13) | |
| Valine, leucine and isoleucine degradation | (12) | |
| Glycine, serine and threonine metabolism | (12) | |
| Starch and sucrose metabolism | (12) | |
| Fructose and mannose metabolism | (12) | |
| Tyrosine metabolism | (12) | |
| Glycerolipid metabolism | (12) | |
| beta-Alanine metabolism | (11) | |
| Propanoate metabolism | (10) | |
| Glyoxylate and dicarboxylate metabolism | (10) | |
| Pyruvate metabolism | (9) | |
| Citrate cycle (TCA cycle) | (9) | |
| Drug metabolism, other enzymes | (8) | |
| Terpenoid backbone biosynthesis: (Cholesterol) Homo sapiens (human) | (8) | |
| Pentose phosphate pathway | (8) | |
| Nicotinate and nicotinamide metabolism | (8) | |
| Alanine, aspartate and glutamate metabolism | (8) | |
| Metabolism of xenobiotics by cytochrome P450 | (8) | |
| Histidine metabolism | (7) | |
| Butanoate metabolism | (7) | |
| Cysteine and methionine metabolism | (7) | |
| Drug metabolism: cytochrome P450 | (7) | |
| Steroid hormone biosynthesis | (7) | |
| Aminoacyl-tRNA biosynthesis | (7) | |
| N = 6: One carbon pool by folate | ||
| Biosynthesis of unsaturated fatty acids | ||
| Linoleic acid metabolism | ||
| Lysine degradation | ||
| Ether lipid metabolism | ||
| N = 5: Sphingolipid metabolism | ||
| N-Glycan biosynthesis | ||
| Porphyrin and chlorophyll metabolism | ||
| N = 4: alpha-Linolenic acid metabolism | ||
| Phenylalanine metabolism | ||
| Retinol metabolism | ||
| Synthesis and degradation of ketone bodies | ||
| Fatty acid elongation | ||
| Butirosin and neomycin biosynthesis | ||
| Glycosaminoglycan degradation | ||
| Steroid biosynthesis | ||
| N = 3: Glycosaminoglycan biosynthesis: chondroitin sulfate | ||
| Pantothenate and CoA biosynthesis | ||
| Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | ||
| Mucin type O-Glycan biosynthesis | ||
| Pentose and glucuronate interconversions | ||
| Selenocompound metabolism | ||
| Ascorbate and aldarate metabolism | ||
| D-Glutamine and D-glutamate metabolism | ||
| N = 2: Vitamin B6 metabolism | ||
| Riboflavin metabolism | ||
| Cyanoamino acid metabolism | ||
| Glycosaminoglycan biosynthesis: heparan sulfate | ||
| D-Arginine and D-ornithine metabolism | ||
| Glycosphingolipid biosynthesis: ganglio series | ||
| Folate biosynthesis | ||
| Other types of O-glycan biosynthesis | ||
| Caffeine metabolism | ||
| Other glycan degradation | ||
| Sulfur metabolism | ||
| Glycosphingolipid biosynthesis: globo series | ||
| N = 1: Fatty acid biosynthesis | ||
| Sulfur relay system Taurine and hypotaurine metabolism Lipoic acid metabolism Phenylalanine, tyrosine and tryptophan biosynthesis Lysine biosynthesis | ||
| Ubiquinone and other terpenoid-quinone biosynthesis Glycosphingolipid biosynthesis: lacto and neolacto series: Glycosaminoglycan | ||
| Biosynthesis: keratan sulfate | ||
| Neuronal | ||
| Neuroactive ligand-receptor interaction | (42) | |
| Dopaminergic synapse | (39) | 0.00284 |
| Neurotrophin signalling pathway | (35) | 0.00189 |
| Serotonergic synapse | (31) | |
| Glutamatergic synapse | (30) | |
| Cholinergic synapse | (28) | |
| Amphetamine addiction | (27) | 0.000172 |
| Retrograde endocannabinoid signalling | (25) | |
| Axon guidance | (24) | |
| Cocaine addiction | (20) | 0.000885 |
| Long-term potentiation | (19) | |
| Morphine addiction | (18) | |
| GABAergic synapse | (16) | |
| Long-term depression | (15) | |
| Synaptic vesicle cycle | (10) | |
| Nicotine addiction | (2) | |
| Phototransduction | (3) | |
As might be expected, a high proportion of genes are involved in the immune system and in pathogen defence pathways. Many are also involved in the life cycle pathways of a number of viruses, bacteria, and other parasites (Table 1). These stem in part from the common immune and defence mechanisms not only related to the pathogens (chemokine and cytokine activation, etc.), but also related to common signalling networks. The involvement of dedicated bacterial and viral defence pathways in the interactome (NOD, RIG1, and cytosolic DNA-sensing pathways) is likely to impact upon viral defence, although in which direction is impossible to determine. Interestingly, T. gondii produces an interferon-like substance with antiviral activity [50]. The host intestinal microbiome also influences T. gondii and is also able to act as an adjuvant in response to T. gondii infection by stimulating dendritic cells that provide the immunostimulation necessary to combat the parasite [51]. Such effects and the shared pathways between pathogens highlight an important potential cross talk between elements of the microbiome.
Diverse pathogens are implicated in all of the diseases in this study, and many of the pathways traced out by the disease susceptibility genes, per se, (posted on the PolygenicPathways website) also involve multiple viral and pathogen life cycle and immune-related pathways.
A number of cancer-related pathways are highly represented in the T. gondii interactome (Table 1). While a recent study has suggested its involvement in brain cancer, based on a correlation between cancer mortality and T. gondii seroprevalence [52], the parasite is able to arrest the growth of other cancerous cells via stimulation of the immune response and inhibition of angiogenesis. Antitumour effects have been observed in relation to spontaneous mammary tumours, leukaemia, lung cancer, and carcinogen-induced tumours following injections of Toxoplasma antigen or viable parasites in laboratory animals or cells [53].
Several autoimmune and atopic disease networks are involved in the parasite interactome. A high T. gondii antibody seroprevalence (as well as to the cytomegalovirus and the Epstein-Barr virus) has been observed in systemic lupus erythematosus, and it has been suggested that antibodies raised to the pathogen may contribute to the autoimmunity characteristic of this condition via pathogen/host protein mimicry [16, 54, 55]. Conversely, T. gondii infection has been shown to prevent the development of lupus-related nephritis in rabbits [56], a factor perhaps related to the immunosuppressant properties of parasitic infection. Toxoplasmosis has been reported to decrease leukocyte, natural killer cell, and monocyte counts in men, while increasing the same in women, with reduced B-cell counts in both [57]. No references were found for relationships between toxoplasmosis and Type 1 diabetes, a pathway also figuring in the interactome. Prior T. gondii infection has been associated with poor outcome in heart transplant patients (allograft rejection) [58]. Toxoplasmosis and other infectious agents have also been linked to cardiac myopathy [59–62], and diverse pathways of which were concentrated in the T. gondii interactome. In relation to asthma, the hygiene hypothesis, linking a reduced incidence of childhood infections (in general) to the worldwide increase in asthma and other allergic conditions, may be related to the concentration of T. gondii interactome genes within the asthma pathway, although a positive correlation of T. gondii infection and asthma has also been noted in Sweden [63–65]. The parasite clearly has multiple effects on diverse immune-related networks as noted above, and such effects are likely to be both beneficial and nefarious. For example parasite-related immunosuppression may well be useful (but perhaps not advisable) in autoimmune diseases such as multiple sclerosis but might also be expected to favour other infections.
Many of the more specific signalling networks within the interactome (Table 2) can be related to the general processes described above. While the MAP kinase pathway is involved in a multitude of functions, the JAK/STAT pathway is involved in cytokine signalling, also bridging cytokine activation to cancer pathways [66]. The calcium signalling pathway is also activated by many processes and more specifically by voltage or receptor-gated ion channels (and is relevant to the “channelopathies” implicated in autism, depression, bipolar disorder and schizophrenia, and in neurological disorders [67, 68]) or by processes modulating intracellular stores, while the phosphatidylinositol signalling system is also involved in the actions of multiple messengers. TGF beta regulates proliferation, apoptosis, differentiation, and migration (definition from KEGG). Calcium channel blockers, calmodulin antagonism, or extracellular calcium depletion diminish cellular invasion by the parasite [69, 70]. The P53 and growth factor signalling networks (ErbB, VEGF) can be cancer related, while insulin signalling is evidently related to diabetes. PPAR receptors control the transcription of many genes especially those related to fatty acid metabolism, but also those involved in cell proliferation and differentiation [71]. These and other pathways control a host of processes from embryonic differentiation to cellular death and apoptosis, and many metabolic pathways that are too numerous to individually review.
In relation to the diseases that are the object of this study, the Alzheimer’s and Parkinson’s disease pathways were both represented, as were the complement, PPAR, and terpenoid (cholesterol synthesis) pathways relevant to Alzheimer’s disease [72], and the ubiquitin pathway relevant to Parkinson’s disease and other degenerative disorders [73]. Erbb signalling is highly relevant to the control of peripheral and central myelination [74], and thus to multiple sclerosis and Alzheimer’s disease, but also to a range of psychiatric disorders including autism, anorexia, ADHD, bipolar disorder, depression, and schizophrenia [75]. Myelin is exquisitely sensitive to oxidative stress and glutathione depletion (c.f. glutathione pathways), and the glutathione precursor N-acetylcysteine has been shown to be of benefit in a number of psychiatric disorders [76–80]. The diverse neurotransmitter pathways and many signalling networks are also relevant to most of these conditions. Rather than single out any particular pathway from this extensive dataset (Tables 1 and 2), suffice it to say that parasitic infection has massive effects upon a variety of host signalling networks, metabolic pathways, and processes. These are nevertheless relatively selective, in the sense that certain pathways are more affected than others. In addition, within each disease dataset, the spectrum of pathways within the overlapping datasets is distinct and biologically relevant, as detailed below.
4.2. Enrichment of Interactome Genes within Susceptibility Gene Datasets (Table 3)
| Disease | N Genes | % involved in T. gondii interactome | Condition | Observed | Expected | Enrichment (fold) | Mean enrichment (A + B)/2 | P value |
|---|---|---|---|---|---|---|---|---|
| Multiple Sclerosis | 408 | 32.5 | Susceptibility genes in interactome (A) | 135 | 54.6 | 2.47 | 2.83 | 1.22E − 71 |
| Interactome genes in disease dataset (B) | 135 | 42.4 | 3.18 | |||||
| Alzheimer′s | 432 | 27.3 | Susceptibility genes in interactome | 118 | 57.8 | 2.04 | 2.33 | 2.26E − 41 |
| Interactome genes in disease dataset | 118 | 44.9 | 2.63 | |||||
| Schizophrenia | 759 | 21.1 | Susceptibility genes in interactome | 160 | 101.6 | 1.57 | 1.80 | 3.06E − 27 |
| Interactome genes in disease dataset | 160 | 78.9 | 2.03 | |||||
| Bipolar disorder | 443 | 21.2 | Susceptibility genes in interactome | 94 | 59.3 | 1.58 | 1.81 | 5.36E − 17 |
| Interactome genes in disease dataset | 94 | 46.05 | 2.04 | |||||
| Depression | 221 | 23.5 | Susceptibility genes in interactome | 52 | 29.6 | 1.76 | 2.01 | 2.41E − 13 |
| Interactome genes in disease dataset | 52 | 22.97 | 2.26 | |||||
| Childhood obesity | 73 | 31.5 | Susceptibility genes in interactome | 23 | 9.77 | 2.35 | 2.69 | 2.32E − 12 |
| Interactome genes in disease dataset | 23 | 7.58 | 3.03 | |||||
| Parkinson′s disease | 263 | 19.7 | Susceptibility genes in interactome | 52 | 35.21 | 1.47 | 1.69 | 3.82E − 08 |
| Interactome genes in disease dataset | 52 | 27.34 | 1.90 | |||||
| ADHD | 237 | 17.7 | Susceptibility genes in interactome | 42 | 31.73 | 1.32 | 1.51 | 8.01E − 05 |
| Interactome genes in disease dataset | 42 | 24.63 | 1.70 | |||||
| Autism | 1117 | 12.7 | Susceptibility genes in interactome | 142 | 149.55 | 0.95 | 1.08 | 0.013 |
| Interactome genes in disease dataset | 142 | 116.13 | 1.22 | |||||
| Anorexia | 74 | 16.2 | Susceptibility genes in interactome | 12 | 9.91 | 1.21 | 1.38 | 0.09 |
| Interactome genes in disease dataset | 12 | 7.69 | 1.55 | |||||
| Chronic Fatigue | 95 | 12.6 | Susceptibility genes in interactome | 12 | 12.72 | 0.94 | 1.08 | 0.48 |
| Interactome genes in disease dataset | 12 | 9.87 | 1.21 |
T. gondii interactome genes were significantly enriched in the susceptibly gene datasets for all diseases with the exception of anorexia and chronic fatigue and represented from ~13% (autism) to 33% (multiple sclerosis) of the total number of susceptibility genes analysed, with enrichment values from 1.08 to 2.83 fold the expected number (Table 3). For schizophrenia, the fold enrichment (interactome genes in susceptibility gene dataset) of 2.03 compares with a recent meta-analysis of T. gondii seroprevalence studies providing an odds ratio (OR) of 2.71 [81]. A further meta-analysis showed significant associations of schizophrenia with infections by human herpesvirus 2 (OR = 1.34), Borna Disease Virus (OR = 2.03), human endogenous retrovirus W (OR = 19.31), Chlamydophila pneumoniae (OR = 6.34), and Chlamydophila psittaci (OR = 29.05), including values far in excess of those for any gene [82]. For schizophrenia at least, these data and ample evidence from epidemiological and animal behaviour studies [83–85] firmly advocate toxoplasmosis as a significant cause of the disease, in those with a particular genetic constitution. The ability of the parasite to manipulate dopaminergic metabolism (via its own tyrosine hydroxylase) [86] and the involvement of NMDA receptor (e.g., glutamatergic signalling and long-term potentiation), serotonin, or cannabinoid-related signalling networks within the interactome is relevant to the drug-induced psychosis associated with the amphetamines, LSD, cannabis, or phencyclidine (see [87]). Dopamine also increases the number of T. gondii tachyzoites in cultured fibroblasts suggesting that neurotransmitters may also be able to manipulate the parasite [88].
For each disease, and across diseases, the types of susceptibility genes influenced were distinct and relatively selective for each disease. This was assessed in two ways: firstly by statistical analysis of the enrichment of KEGG pathways in each overlapping T. gondii interactome/disease dataset and secondly by a comparison of individual shared and specific overlapping interactome/disease genes across four diseases (the maximum possible using the Venny tool). The diseases analysed in this way were Alzheimer’s disease and multiple sclerosis, bipolar disorder, and schizophrenia.
4.3. Overlapping Interactome/Susceptibility Genes Common and Specific to Four Diseases (Table 4, Figure 1)
| Alzheimer’s | Bipolar | Schizophrenia | Multiple sclerosis | |
|---|---|---|---|---|
| Common to all | APOE GSK3B SYN3 Cytokine IL10 IL1B IL1RN IL6 TNF Oxidative stress GSTM1 ND4 | |||
| Alz, Bip, Sz | Neuronal development/growth DPYSL2 Oxidative stress MAOA NOS1 SOD2 | Neuronal development/growth DPYSL2 Oxidative stress MAOA NOS1 SOD2 | Neuronal development/growth DPYSL2 Oxidative stress MAOA NOS1 SOD2 | |
| Alz, Bip and MS | Chemokine CCL2 Oxidative stress ND1 | Chemokine CCL2 Oxidative stress ND1 | CCL2 ND1 | |
| Bip, Sz and MS | Immune CTLA4 IFNG Other MMP9 PDE4B | Immune CTLA4 IFNG Other MMP9 PDE4B | Immune CTLA4 IFNG Other MMP9 PDE4B | |
| Alz and Ms | Immune CCL3 CCR2 CD14 CD86 IL8 TAP2 TGFB1 Oxidative stress GSTM3 NOS2 Other APOC2 FAS GRN ICAM1 SERPINE1 TOMM40 | APOC2 CCL3 CCR2 CD14 CD86 FAS GRN GSTM3 ICAM1 IL8 NOS2 SERPINE1 TAP2 TGFB1 TOMM40 | ||
| Alz, Sz and MS | Immune/inflammation C4A PTGS2 Oxidative stress ATP6 CYTB Other CAV1 ESR1 MMP3 PPARG VDR | ATP6 C4A CAV1 CYTB ESR1 MMP3 PPARG PTGS2 VDR | ATP6 C4A CAV1 CYTB ESR1 MMP3 PPARG PTGS2 VDR | |
| Bip and Sz |
|
|
||
| Alz and Sz |
|
ABCA1 C4B EBF3 IL18 IL1A KLF5 LPL PCK1 | ||
| Alz and Bip | HSPA5 Growth IGF1 | HSPA5 Growth IGF1 | ||
| MS and SZ | Immune CCR5 CD4 CNTF HLA-A IGH@ IL12B IL2 IL4 LTA Other MYH9 PRKCA UCP2 | Immune CCR5 CD4 CNTF HLA-A IGH@ IL12B IL2 IL4 LTA Other MYH9 PRKCA UCP2 | ||
|
|
|
|
|
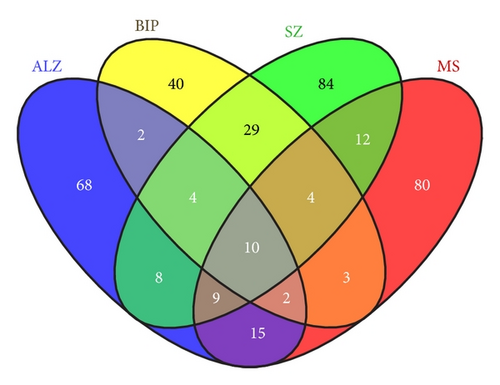
The permutations of genes common or specific to the various chosen diseases (Alzheimer’s disease, bipolar disorder, schizophrenia, and multiple sclerosis) are shown by the Venn diagram Figure 1 summarised in Table 4. All of these genes are members of the host/pathogen interactome. Several immune/cytokine and oxidative stress related genes, with different identities, but similar roles, appear as common risk factors across various permutations of diseases, which are all characterised by immune activation [89–91] and oxidative stress [92, 93].
Bipolar disorder and schizophrenia share many common genes, risk factors, endophenotypes, and subpathologies, and interactome genes relevant to certain of these are related to circadian rhythm, dopaminergic and glutamatergic neurotransmission, growth factors, and signalling networks as highlighted in previous reviews [75, 94, 95].
After sifting through these common subsets, the overlapping T. gondii interactome/susceptibility genes specific to each disease are remarkably relevant to the key primary pathologies in each. They include APP processing, cholesterol and lipoprotein function, complement and immune related genes, and oxidative stress, apoptosis and ubiquitin genes in Alzheimer’s disease [96–100]. In bipolar disorder, monoamine/GABA, signalling, adhesion, and ion transport genes are highlighted (see above and [101–103]) while in schizophrenia, monoamine/glutamate/neuregulin neuronal development and associated signalling related genes figure prominently, along with those related to adhesion, oxidative stress, and immune activation (see above). In multiple sclerosis, almost the entire common dataset is related to immune function and associated signalling pathways, that are relevant to the autoimmune aspects of the disease [104, 105], with a limited number of genes related to oxidative stress and apoptosis.
While the evidence for an involvement of toxoplasmosis in psychiatric disorders is relatively strong, there is less work either in the human condition or in animal models in the case of neurological disorders, such as Alzheimer’s or Parkinson’s diseases or multiple sclerosis. Toxoplasmosis has, however, been associated with a loss of grey matter density in schizophrenic patients, but not in controls, suggesting an influence on degenerative components [8]. T. gondii infection may not always be deleterious. For example, it inhibits the development of arthritis in mice deficient in the interleukin receptor antagonist (IL1RN) [106]. T. gondii infection is also able to reduce infarct size in focal cerebral ischaemia in mice, an effect attributed to the ability of infection to increase the expression of nerve growth factor, as well as that of anti-inflammatory cytokines and of glutathione and oxidative stress protective genes, while reducing the expression of proinflammatory cytokines [107].
Parasites have learnt to live with us for many millennia, and their immunosuppressant effects appear to be a relatively common defence mechanism. Indeed, the use of helminths (parasitic worms) has been suggested in a number of autoimmune settings including irritable bowel disease and multiple sclerosis [108]. A clinical trial with helminth egg infection (Trichuris Suis Ova) in autism is also listed at http://clinicaltrials.gov/, based on anecdotal reports of effectiveness in relation to certain symptoms. The preponderance of immune related host/pathogen genes in the multiple sclerosis dataset (and to a lesser extent within other datasets) may be related to these potentially beneficial effects, although the clinical use of T. gondii would be contraindicated by its malevolence directed elsewhere.
4.4. KEGG Pathway Analysis of the Overlapping Datasets Specific to Each Disease (Tables 5 and 6)
| Immune and defence | Diseases | Other infections | |
|---|---|---|---|
| ADHD | None | None | None |
| Autism |
|
|
Leishmaniasis 0.00605 |
| Anorexia | None | None | None |
| Childhood obesity |
|
|
|
| Depression |
|
|
|
|
|
|
|
|
|||
| Bipolar disorder |
|
|
|
| Schizophrenia |
|
|
|
|
|
|
|
|
|
|
|
| Multiple sclerosis |
|
|
Chagas disease (American trypanosomiasis) 5.89E − 22 Influenza A 2.81E − 19 Toxoplasmosis 4.01E − 18 |
|
|
||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Alzheimer’s |
|
|
|
|
|
|
|
| Parkinson’s |
|
|
|
| Signalling networks | Process | Metabolism | Neuronal | |
|---|---|---|---|---|
| ADHD | Calcium signalling pathway 0.00442 |
|
|
|
| Autism | VEGF signalling pathway |
|
None |
|
| Anorexia | None | None | None |
|
| Childhood obesity |
|
None | Glycerolipid metabolism 0.00525 | None |
| Depression |
|
|
|
|
| Bipolar disorder |
|
Apoptosis 0.0000464 |
|
|
| Schizophrenia |
|
|
Glutathione metabolism 0.00583 |
|
| Multiple sclerosis |
|
|
||
| Alzheimer’s | PPAR signalling pathway 0.0000000416 | Osteoclast differentiation 0.000351 |
|
|
| Parkinson/s | MAPK signalling pathway 0.00623 |
|
|
|
4.4.1. Immune and Pathogen Defence Pathways Common to Most Diseases
The KEGG pathways influenced by T. gondii (restricted to the overlapping interactome genes within each disease dataset) are posted at http://www.polygenicpathways.co.uk/toxoplasmosis.htm, and the CPDB enrichment analysis depicted in Tables 5 and 6. These tables report only the significantly enriched pathways, but many others figure within these overlapping datasets. In all diseases, except for ADHD and anorexia, the significantly enriched subsets involved immune or defence related pathways. For the most part (autism, childhood obesity, depression, bipolar disorder and schizophrenia, Alzheimer’s and Parkinson’s disease, and multiple sclerosis), the bacterial defence NOD signalling network was involved, while the similar Toll pathway was more restricted (Alzheimer’s and Parkinson’s disease and multiple sclerosis, bipolar disorder, and schizophrenia). The RIG1 and cytosolic DNA-sensing pathways recognise viral nucleic acids. The RIG-1 pathway was significantly enriched in multiple sclerosis and schizophrenia, while the cytosolic DNA-sensing pathway was enriched in Alzheimer’s and Parkinson’s disease as well as in multiple sclerosis and schizophrenia. Diverse pathogen life cycle pathways were enriched in all but the ADHD and anorexia datasets.
4.4.2. Childhood Obesity and Anorexia
There are few studies relating either obesity or anorexia to toxoplasmosis in man, although both anorexia or subsequent partial weight gain postinfection, as well as hypermetabolism have been associated with T. gondii infection in laboratory and farm animals [109–111].
The only significantly enriched pathways common to the T. gondii interactome in anorexia all relate to neuronal systems (dopamine, serotonin, and addiction pathways).
In childhood obesity, a number of autoimmune related pathways were highlighted, as well as the Alzheimer’s disease pathway, pathways related to PPAR signalling (regulating fatty acid metabolism), and glycerolipid metabolism. A recent review has highlighted the risk promoting effects of midlife obesity (and several other preventable risk factors) in relation to Alzheimer’s disease [112]. The childhood obesity epidemic, fuelled largely by dietary and sedentary culture [113], has been associated with an increased risk of affective disorders in adulthood [114] and has also led to an increased incidence of a number of diseases in young children (dyslipidemia, carotid artery atherosclerosis, cardiac problems, hypertension, the metabolic syndrome, and diabetes and fatty liver disease) [115–118] that were previously the reserve of old age. Many of these are also risk factors for Alzheimer’s disease and are able, per se, to increase cerebral beta-amyloid deposition in laboratory models, perhaps a herald for the unwelcome imminence of dementia in young adults.
Diet, including saturated fat [119, 120], affects the microbiome, and a recent study has shown that, in infants fed formula or breast milk, changes in the gut microbiome can alter the expression of genes related to the innate immune system [121]. This microbiome/immune link may be important in the development of inflammation and metabolic diseases [120]. There do not appear to have been any microbiome studies in relation to T. gondii. However, the parasite scavenges host cholesterol, while host fatty acids and low-density lipoproteins stimulate a T. gondii acyl-CoA, cholesterol acyltransferase, which then provides cholesteryl esters that the parasite needs for its survival [122]. Fatty diets would certainly be expected to impact upon the success of this parasite, which in turn must influence the lipid metabolism of the host. Indeed, T. gondii infection may even possess beneficial effects in hypercholesterolaemic conditions in mice, reducing the development of atherosclerosis via cholesterol and lipoprotein scavenging effects [123].
Pathway correlates such as these of course predict relationships but not directionality, which can only be imputed by prior knowledge and future research. Certain of the pathways common to the T. gondii interactome and obesity (and to Alzheimer’s disease, see below) could well reflect a beneficial component of parasitic infection.
4.4.3. Attention Deficit Hyperactivity Disorder and Autism
No clinical studies have specifically linked ADHD or autism to toxoplasmosis, although hyperactivity, modified social interactivity, and sensorimotor effects are features of infection in mice that are of relevance to both conditions [124–126].
In ADHD, the primary common emphasis was on the calcium signalling pathway to a number of metabolic pathways: phenylalanine and tyrosine (DDC and MAOA), tyrosine, histidine (DDC, HNMT, and MAOA) tryptophan (ACAT1, DDC, and MAOA), and unsaturated fatty acid synthesis (FADS1 and FADS2) and to neurotransmitter pathways (cocaine addiction and ligand/receptor interactions). This is a relatively small dataset, but it highlights an important distinction for bacteria or parasites, which, unlike viruses, participate in substrate and metabolite exchange with the host, enabling a much greater effect on metabolic pathways. This influence may be particularly relevant to the reported risks and benefits of various types of diets in many diseases, and in particular, saturated and unsaturated fats [127].
In autism, various cardiomyopathy pathways were enriched in the overlapping dataset. Autistic components have been observed in a number of cardiomyopathy disorders (MELAS and Timothy syndromes and Danon disease) [128–130]. T. gondii seropositivity has also been associated with cardiomyopathy [60]. Cellular adhesion and the extracellular matrix play a key role in brain development and in autism [131, 132], and these pathways were the only significantly enriched “processes” in the overlapping dataset. VEGF signalling, dopamine, serotonin, and addiction pathways, but no metabolic pathways, were also enriched. Serum VEGF levels have been reported to be reduced in severely affected autism cases [133].
4.4.4. Depression and Bipolar Disorder
Although perhaps less evident than with schizophrenia, toxoplasmosis has nevertheless been associated with prenatal depression, depression, bipolar disorder, and with a history of suicide attempts in recurrent mood disorders [9, 10, 134, 135].
In depression, as well as immune, defence, and diverse pathogen related pathways, autoimmune diseases, hypertrophic cardiomyopathy, rheumatoid arthritis and osteoclast differentiation, cancer pathways, and Alzheimer’s disease were overrepresented in the overlapping dataset. Depression and arthritis have been reported as comorbid conditions [136], and prior depression is a significant risk factor in both cardiac conditions and Alzheimer’s disease [137, 138]. Numerous studies have implicated the VEGF pathway is relevant to depression and to the mechanism of action of antidepressants [139]. With regard to transforming growth factor, TBF-beta, an anti-inflammatory cytokine, an imbalance of pro- and anti-inflammatory cytokines has been observed in major depression studies [140]. Neuronal pathways primarily concerned reward/addiction, glutamate, dopamine, serotonin, and cannabinoid networks. An overrepresentation of phenylalanine and tryptophan metabolism is also relevant. The circadian clock pathway, which was also over-represented, plays a key role in depression and related disorders [141]. In drosophila, the circadian clock regulates the phagocytosis of bacteria [142], and within its many functions are the control of the immune system [143]. Unsaturated fatty acid metabolism again figured in this group, and the general benefits of modifying saturated/unsaturated fat ratios in diet are increasingly recognised, including in the area of psychiatry [144].
The overlapping dataset in bipolar disorder also concerned immune related pathways, several autoimmune disease networks (Type 1 diabetes, arthritis (and osteoclast differentiation), graft-versus-host disease, and allograft rejection), and a number of pathogen life cycle pathways. In relation to cancer pathways, slight increases in overall cancer risk have been reported in both bipolar disorder and schizophrenia, which appear to be gender dependent [145]. In relation to the Alzheimer’s disease pathway (which independently figures in all KEGG pathways related to susceptibility genes alone in most of these disorders), prior psychiatric illness has been shown to be generally associated with an increased risk of developing dementia [146]. Common pathological features across many psychiatric disorders and Alzheimer’s disease also include white matter changes related to demyelination [147, 148]. Many of the stressors involved in these conditions (starvation, viruses, infections and fever, cytokines, oxidative, and endoplasmic reticulum stress) converge on a pathway that ultimately inhibits translation initiation and protein synthesis. This network is counterbalanced by growth factors and neurotransmitter influences that affect plasticity and growth and is particularly important in regulating oligodendrocyte viability, myelination, and synaptic plasticity [149] (c.f. the neurotrophin pathway within this dataset and related glutamatergic and growth factor signalling networks in others). Neurotransmitter networks within the overlapping bipolar/interactome are predominantly related to dopamine and reward pathways and to tyrosine, phenylalanine, and tryptophan metabolism.
4.4.5. Schizophrenia
The link between schizophrenia and toxoplasmosis is perhaps the strongest in relation to published studies [6, 82, 150–152], and of particular relevance is the parasite’s ability to increase cerebral dopamine levels (see above). In this respect, the overlapping interactome/gene dataset was enriched in dopaminergic pathways, and also in those related to serotonergic and glutamatergic transmission as well as cocaine and amphetamine addiction. As in most cases autoimmune and atopic diseases, which are commonly associated with schizophrenia, were well represented. In many autoimmune conditions the link with schizophrenia was positive and gender specific, while an inverse association between schizophrenia and rheumatoid arthritis was observed [153, 154] (c.f. the concentration of osteoclast differentiation pathways in this dataset). Gluten sensitivity (characterised by antibodies to a gluten constituent protein, gliadin), other food antibodies, and celiac disease have also been associated with schizophrenia. Food antigens in schizophrenic patients have been shown to be correlated to the presence of T. gondii antibodies. Interestingly, the antiparasitic agent artemisinin reduces the titre of antibodies to gliadin in a subset of schizophrenic patients, and these observations testify to the ability of the parasite to modulate immune function (and perhaps the antigenicity of other proteins). However, artemisinin did not reduce the titre of antibodies to T. gondii, nor did artemisinin (as add on therapy) have significant effects on symptomatology [155–157]. Artemisinin and its analogues are known to produce neurotoxic effects in laboratory models, an effect possibly linked to excitotoxicity and oxidative stress [158, 159], and clearly more suitable agents are needed in the research domain. The overlapping dataset also included significant enrichment of adhesion molecule, glutathione, and growth factor and related signalling pathways (VEGF, MAPK, and Wnt, but not ERBB signalling, although this pathway is affected by T. gondii). The PPAR network was also enriched in this dataset and is relevant in relation to the inflammatory arm of this pathway and to the ability of the pathway to regulate cholinergic and dopaminergic function [160, 161]. With regard to the cancer pathways in this dataset, schizophrenia has been associated with a reduced cancer incidence, but with no familial explanation, suggesting a nongenetic reason that may conceivably be related to the abilities of T. gondii, and other relevant pathogens, to favour the promulgation of one disease, but perhaps protect against another. In relation to the overlapping Alzheimer’s disease pathway within this dataset, the association of prior psychiatric illness with dementia has already been mentioned [146].
4.5. Neurodegenerative Disorders
4.5.1. Parkinson’s Disease
There are only limited human seroprevalence studies and no apparent animal studies specifically in relation to the substantia nigra, linking toxoplasmosis to Parkinson’s disease [11, 12]. Nevertheless the overlap between the T. gondii interactome and susceptibility genes figures certain key pathways that may merit further research.
The interactome/genetic overlap for significantly enriched pathways in Parkinson’s disease in relation to neurotransmission was restricted to dopaminergic systems, and a number of key genes including those of the mitochondrial respiratory chain (ATP6, CYTB, and ND2), the quinone reductase NQO2, and two key Parkinson’s disease genes (PINK1 and UCHL1) figure within the enriched T. gondii interactome. While an ability of T. gondii to promote dopamine synthesis might be considered beneficial in Parkinson’s disease, it has also been shown that dopamine promotes synuclein conformational changes, which may directly contribute to pathology [162]. As with other diseases, autoimmune networks, cancer pathways, and Alzheimer’s disease were represented. Cancer and neurodegenerative diseases in general appear to be inversely correlated [163].
4.5.2. Alzheimer’s Disease
Any link between Alzheimer’s disease and toxoplasmosis is limited to a seroprevalence study [13] and to scattered case reports [164, 165].
In Alzheimer’s disease, the significantly enriched pathways included PPAR signalling, terpenoid biosynthesis (cholesterol synthesis) concerned with fatty acid, lipid, and cholesterol homoeostasis, and the arginine and proline metabolism pathway, primarily concerning nitric oxide, all of which play a key role in Alzheimer’s disease physiology [166, 167].
Several pathogens (herpes simplex, C. pneumoniae, treponemas, and spirochetes) [24, 168, 169] increase beta-amyloid deposition. The gamma secretase network and APP are localised in immunocompetent dendritic cells, and, as the amyloid peptide possesses antimicrobial and antiviral effects [170, 171], beta-amyloid production may well be a general defensive response to pathogen invasion [20]. In normal conditions, it is not known whether beta-amyloid production is also a response to larger parasites, or whether beta-amyloid has antiparasitic activity.
In Tg2576 transgenic mice (the Swedish APP mutation), T. gondii infection in fact reduces cerebral beta-amyloid deposition and increases the levels of anti-inflammatory cytokines, effects attributed to the immunosuppressant effects of infection [172]. In relation to the cholesterol related genes in the Alzheimer’s disease T. gondii dataset, the parasite cannot synthesis its own sterols and scavenges host cholesterol. Its growth in macrophages can be inhibited by statins [173]. While a living cholesterol lowering agent might be considered useful in the periphery, such effects may be deleterious if limited to cerebral areas, as the brain synthesises its own cholesterol. This is mostly present in myelin and is generally indispensable for function [167]. In the Alzheimer’s disease Tg2576 transgenic model, T. gondii lysate antigen inhibits the production of nitrites in microglial cells, contributing to the protective effects of infection in this model [172]. As with obesity, certain interactome/susceptibility gene pathways involved in parasitic infection might well be considered as beneficial.
4.5.3. Multiple Sclerosis
Although by far the most enriched dataset in terms of interactome/susceptibility gene overlaps, there appear to have been no studies either in the clinic or in relation to myelination in laboratory studies linking multiple sclerosis and toxoplasmosis. A study in 3 pairs of identical twins reared apart was generally inconclusive, although T. gondii or other pathogen seropositivity were observed in some cases [174]. Further work will be of interest in relation to this close association.
In multiple sclerosis, the major overlapping pathways primarily concerned cytokine and TGF-beta signaling, the related JAK-STAT pathway, and the ErbB and p53 signalling pathways that plays a key role in myelination [175, 176].
5. Summary
Within each disease dataset, the susceptibility genes that overlap with the T. gondii interactome, analysed by either method, appear highly relevant to the pathological processes and physiology of the disease. This convergence suggests a massive effect of infection on numerous processes. However, while some may be deleterious, (e.g., the promotion of dopaminergic activity in relation to psychosis), others may be beneficial (e.g., immunosuppression in autoimmune diseases). Even within any particular disease, the diverse effects of the parasite could be either favour or inhibit the development of particular endophenotypes. As suggested below, the overall direction taken and the resulting pathology are likely to depend upon a combination of factors including the strain of parasite, the timing and localisation of infection, our prior immune status, and the susceptibility genes.
In many cases, the signalling networks influenced by susceptibility genes either per se or within the overlapping host/pathogen interactome involve many diseases other than the primary disease concerned. Diseases are often associated with other diseases, either positively or inversely [177]. For example degenerative disorders may be inversely associated with cancer [163, 178]. This may be related to particular signalling networks, for example growth factor signalling pathways are essential for myelination or involved in long-term potentiation, but excessive stimulation will promote cancerous growth. The ability of T. gondii (and other pathogens) to affect so many processes, which may be either deleterious or beneficial to various disease-related networks, suggests that pathogens may also be the pivot around which such relationships revolve.
5.1. Autoimmunity and Host/Pathogen Protein Homology
Several studies have recently shown that the entire human proteome contains short sequences (pentapeptides to heptapeptides or longer gapped consensi) that are identical to those within proteins expressed by numerous viruses, bacteria and other pathogens. For diverse pathogens, these human homologues appear to be concentrated within networks that are relevant to diseases in which the pathogen is implicated [35, 37, 38, 179, 180]. This problem is extensive and concerns all human proteins, along their entire length. For example, there are 18,000 pentapeptide overlaps between the poliovirus and the human proteome [181] while a single immunogenic pentapeptide (VGGVV) within the beta-amyloid peptide is identical to that within proteins from the herpes simplex virus and from 68 other viral species [19]. The extensive host/pathogen interactomes of numerous viruses, bacteria, and parasites no doubt result from this homology which enables pathogen proteins to mimic particular motifs within their human counterparts and to compete for their usual binding partners. Such homology must presumably relate to our evolutionary decent from monocellular organisms and to horizontal gene transfer, a process that applies to all living matter [182]. It is now also appreciated that DNA derived from both DNA and RNA viruses (and not only from retroviruses) has been extensively incorporated into the human genome, and it seems likely that this has also played a role in our evolution, and evidently in the generation of this protein homology [182–185]. Host parasite interactions have also contributed to this gene transfer, and genes from the Chagas disease vector, Rhodnius prolixus, have been found within the genomes of its tetrapod hosts [186]. Peptide homology is more extensive than genetic homology, due to the fact that a number of amino acids can be coded for by several triplet DNA codons (6 for arginine leucine and serine, 4 for alanine, glycine, proline, threonine, and valine, 3 for isoleucine, and 2 for asparagine, aspartate, glutamate, glutamine, cysteine, histidine, lysine, and phenylalanine) (see http://en.wikipedia.org/wiki/DNA_codon). These essentially correspond to single nucleotide polymorphisms that do not modify the translated amino acid. For short peptide sequences, numerous different DNA sequences can thus code for identical peptides.
This extensive homology, and more particularly slightly differing rather than identical peptides (which are more likely to be recognised as nonself) [36, 187], may well also contribute to autoimmunity problems that are evident in many diseases. For example, in Alzheimer’s disease, multiple sclerosis, schizophrenia, and AIDS, antigenic regions of several autoantigens particular to each disease are homologous to proteins expressed by the pathogens implicated in the same disease (including T. gondii and schizophrenia) [18–21, 39].
Diseases currently classified as autoimmune include celiac disease, multiple sclerosis, myasthenia gravis, lupus, rheumatoid arthritis, and inter alia (see Medline Plus article at http://www.nlm.nih.gov/medlineplus/ency/article/000816.htm. However, the autoimmune problem appears to be much more extensive than currently appreciated. For example, using a protein array of 9,486 unique human protein antigens, even control blood samples averaged over 1000 autoantibodies, although with extreme intersample variation. As only ~30% of the human proteome was used in this experiment, we may each eventually accumulate over 3000 autoantibodies, irrespective of any particular disease. However, in both Parkinson’s and Alzheimer’s disease the target profile of the autoantibodies is distinct and can be reliably used as a diagnostic and predictive tool [188, 189]. Autoimmune signatures, with diagnostic predictive value have also been reported in multiple sclerosis [190], breast cancer [191], and nonsmall cell lung cancer [192]. Such data, (in diseases generally not regarded as autoimmune) and the recognition that so many diseases are characterised by immune activation and inflammation suggest that further research in this area would be fruitful in relation to the understanding of the pathologies and eventual treatment of many diseases.
The immune system is trained, in early life, not to recognize the body’s own proteins as self [193]. These bioinformatics data suggest that the multiple autoantibodies seen in man (even in the absence of disease) may stem not from some inherent malfunction of the immune system itself, but from antibodies raised to the numerous pathogens that we randomly encounter during the course of our lifetime. Because of this extensive host/pathogen homology, such antibodies are also likely to target human proteins, and even if the pathogen is eliminated, continued encounter of these human homologues would sustain an autoimmune response. In this way, pathogens might be able to influence disease processes, even when no longer present, perhaps accounting for numerous studies that have failed to find pathogen DNA or protein within diseased tissue, a finding often cited as evidence against pathogen involvement, as recently applied to the controversial implication of the XMRV virus with chronic fatigue or prostate cancer [194–197]. The prospect that autoimmunity is pathogen related suggests that such agents may be able to punch far above their weight and influence biological processes even after their successful removal. This entails a revision of Koch’s postulate as already discussed in a recent review on autoimmunity and the metagenome [177]. This autoimmune scenario might also explain why the antiparasitic agent artemisinin failed to influence psychotic symptoms (as add-on therapy) in schizophrenia [156], as destruction of the parasite needs not to affect the behaviour of antibodies raised to it.
Antibodies to pathogens are clearly cross-reactive with cerebral tissue, although the precise targets remain to be identified. For example 14/25 antibodies to 17 neurotropic pathogens, including Borrelia burgdorferi, T. gondii, and various DNA and RNA viruses were found to bind to western blots of human nervous tissue [198]. It is impossible to verify cross-reactivity solely from sequence analysis, but the ability of pathogen antibodies to react with human proteins could perhaps be tested in bulk using the protein arrays described above. It is now known that antibodies can enter cells, transported by the pathogens to which they bind, [199], and are also able to traverse the blood brain barrier [200]. Antibodies to receptors can also enter cells using the receptor endocytosis apparatus [201]. Antibodies can have devastating pathological consequences. For example, in transgenic mice engineered to express nerve growth factor antibodies only in lymphocytes, the blood brain barrier is soon disrupted, with cerebral antibody entry provoking extensive cortical degeneration, cholinergic neuronal loss, tau hyperphosphorylation, and beta-amyloid deposition (i.e., the cardinal pathology of Alzheimer’s disease) [202]. This phenomenon is applicable to human diseases, including Sydenham’s chorea, believed to be caused by streptococcus induced antibodies which cross-react with basal ganglia antigens [203]. The same streptococcal pathogens (and likely a similar mechanism) have been implicated in paediatric autoimmune neuropsychiatric disorders (PANDA’s) whose diverse symptoms include tics, and dystonias, Tourette syndrome, and obsessive-compulsive disorder [204].
If autoantibodies do indeed play a key role in the pathogenesis of many diseases, then it is likely that their removal may be of benefit. However, given the large number of autoantibodies, some of which may well be beneficial and also required for pathogen defence, this may be no easy task. However, the research so far suggests that the number of autoantibodies specific to a particular disease may be more limited, allowing scope for analysis of their pathological or redemptive properties.
5.2. Population Genetics and a Proposed Gene/Environment Interaction Model (Figure 2)
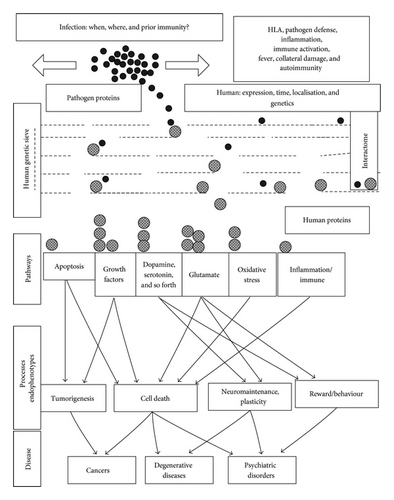
The mechanisms described above provide a general example of multiple gene/environment interactions in relation to a single pathogen interactome, where several thousand genes (human and protozoan) are involved. Even for a simple population genetics model, with two genes, two risk factors, and a single cause, varying permutations can dramatically influence the eventual outcome. For example, the light and dark coloured genes of the peppered moth, or the light and dark colours of the clean or polluted trees on which they alight, can all be either risk promoting or protective depending on the varying permutations (the light gene “kills” the moth alighting on dark trees but is protective on the lighter trees, etc.) [205]. Neither gene, nor risk factor is relevant if there are no hungry birds or at night time. If one splits a complex disease into its component parts and gives the number of interacting processes involved in the T. gondii interactome, even this single pathogen could act either as a cause, a risk promoter, or as a protective agent, depending upon the pathways that it influences the most. For example, its effects on dopamine could promote psychosis or synuclein polymerisation, and its cholesterol scavenging may have beneficial effects in atherosclerosis, but deleterious effects on myelination, immunosuppressive effects might well protect against autoimmunity, but favour other infections, while the host’s inflammatory reaction or associated fever might contribute to inappropriate collateral damage.
These complex interactions are nevertheless based on a relatively simple concept; that each interaction has an effect on the processes and pathways regulated by the human protein concerned. This suggests a model that may have general application to the many other pathogens and environmental agents implicated in these diseases.
If one imagines the T. gondii proteins as a number of spheres, each with particular affinity for certain human genes or proteins, and their human interactome partners as a further series of spheres perched on a genetic ledge whose characteristics and apertures are regulated by polymorphisms, mutations, deletions, translocations, or copy number variations, then the trajectory of each, dropped through this genetic sieve or knocked off the ledge and falling through the apertures, will be influenced both by the strain of pathogen with different host/pathogen affinities, the dropping point, the timing, and localisation of infection, when and where different human genes are expressed and by the polymorphic genes themselves (for both the host and the pathogen).
Each of these human genes controls a particular element of one or many signalling networks, metabolic pathways, structural elements, developmental processes, and so forth, each represented by reception bins at different positions beneath the sieve. Depending upon varying permutations of these factors, the eventual number of spheres in each bin will vary, resulting in a diverse spectrum of pathway disturbance. Each pathway may be affected either positively or negatively, and the eventual assembly of this pathway mosaic leads to particular endophenotypes or subpathologies, which together combine to assemble into a particular disease. In this way, the same pathogen can produce diverse effects ranging from cause to prevention depending on a permutation of circumstance.
The genes, risk factors, and the immune system thus work together to determine the final outcome, while neither per se are likely to provoke a particular disease. While gene/environment interactions are appreciated in both genetic and epidemiological studies, most, particularly in relation to GWAS, are performed without data partitioning in relation to other variables [206]. Many other pathogens (each no doubt with extensive host pathogen interactomes) and many other risk factors are implicated in these and other diseases, and many are able to influence several relevant aspects of pathology (see Section 1). A clearer understanding of these complex effects could perhaps result in a metamorphosis from multiple genes of small effect in large populations to more restricted numbers of greater effect in particular conditions. It is likely that many disease phenotypes have several “causes,” that subsets of overlapping genes are relevant to each, and that despite the mass of data collection and processing entailed, a dissection of these relationships could eventually lead to disease prevention and cure in multiple conditions.
By their very nature, polygenic diseases are complex, with several underlying pathologies and endophenotypes, hundreds of interacting genes, and dozens of environmental risk factors. The failure to replicate either genetic or epidemiological data is a situation peculiar to these diseases, not seen in many other fields. However, the effects of genes and risk factors are clearly conditional and, as illustrated above, may well depend upon each other. Replication inconsistency may well be part of the answer and not part of the problem.
6. Conclusion
The host/pathogen interactome influences ~10% of the human genome products. This may seem a surprisingly high figure, but a similar interactome for the HIV-1 virus, maintained by NCBI http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=DetailsSearch&Term=hiv1interactions%5bproperties%5d, contains 1443 human genes (5.4% of the human genome). Bacteria and larger protozoan parasites, which, unlike viruses, also scavenge for host nutrients, as well as injecting their own metabolites into the host’s environment, thus influence a larger spectrum of biochemical rather than signalling pathways. These data were also collected from experiments using various host (and species) tissues, and it is likely that brain or other tissue or time-specific interactomes would be more selective.
The relevance of many genes to a particular condition is often tested by gene knockout in transgenic models and comforted by the resulting endophenotypes which mimic those of the particular disease [207]. However, risk promoting variants are, for the most part, single nucleotide polymorphisms rather than deletions and while expression may be altered (in either direction) in mRNA or protein expression studies, there is little to suggest a similar knockout in the human condition (see for example the microarray Geoprofiles database at NCBI http://www.ncbi.nlm.nih.gov/sites/geo/).
However, there are two pathogen-related effects that equate to conditional protein knockout which could be cell and regionally and temporally specific. The first relates to the host/pathogen interactome and the second to autoimmunity. If a host protein is engaged with that of a pathogen, it is effectively taken out of circulation during this period, and the pathways in which it is implicated can but be compromised. Secondly, because of extensive homology between pathogen and human proteins, antibody cross reactivity is likely to target the human counterparts of the pathogen antigen, effectively resulting in immunopharmacological knockout. In addition to these knockout effects, immune activation and the general reaction to infection are also likely to influence cellular function, as are the multitude of genes whose mRNA levels are influenced by this and other pathogens. In relation to prenatal effects, laboratory models have shown that maternally administered nonspecific viral DNA mimics and inflammatory agents or cytokines can also induce behavioural disturbances and psychopathology in the offspring [208, 209]. Fever during pregnancy also increases the risk of the offspring later developing autism and schizophrenia [210, 211], and it seems likely that prenatal infection in general is able to markedly affect brain development. The consequences would also depend upon which particular brain process and region is concerned at which period of embryogenesis.
Many other pathogens have been implicated in several of these conditions. In some of the diseases studied, almost one-third of the susceptibility genes were implicated in the T. gondii interactome (Table 3). Other pathogens will also have extensive interactomes, specific to each, but with a degree of overlap, and it would not be implausible if the near totality of susceptibility genes, in certain diseases, were involved in the summated life cycles of these diverse environmental triggers. It would thus seem that many susceptibility genes are related to the causes of disease, rather than (and as well as) to the disease itself. It is likely that stratification of GWAS and other genetic data in relation to infection status and history and many other environmental variables would be useful in determining the contribution of different genes to different risk factors and to their commonly affected pathways.
Many psychiatric disorders are associated with a degree of social stigma and blame often apportioned to the genes, parentage and upbringing, and behaviour of the affected individuals. These and other chronic diseases also place a heavy long-term burden on family, friends, and caregivers [212]. This analysis suggests that T. gondii is a likely cause of certain aspects of psychiatric disorders, but perhaps a protective agent in others. Hopefully, an appreciation that such diseases may well be caused by pathogens and vectored by family pets will help to dispel such prejudice and more importantly create a new framework for the development of new methods of treatment and prevention. Given the massive problem of autoimmunity, however, it may be simplistic to suggest that removing the pathogen will halt the disease, although prevention of its initial access might be expected to affect disease incidence. Such approaches need not necessarily be clinical. For example if toxoplasmosis in cats and other pets was registered as a notifiable disease requiring obligatory treatment by veterinarians, perhaps the incidence of several diseases could be reduced.
Acknowledgments
The author is particularly indebted to the KEGG staff for their pathway contribution and for permission to post pathways on the PolygenicPathways website and to the numerous authors who have provided reprints.




