Communication P Systems on Simplicial Complexes with Applications in Cluster Analysis
Abstract
The purpose of this paper is to propose a new kind of P systems on simplicial complexes. We present the basic discrete Morse structure, membrane structures on complexes, and communication rules. A new grid-based clustering technique is described based on this kind of new P systems. Examples are given to show the effect of the algorithm. The new P systems provide an alternative for traditional membrane computing.
1. Introduction
Membrane computing is a new branch of natural computing which is initiated by Păun et al. at the end of 1998, as an attempt to formulate models from the functioning of living cells [1], just like DNA computing coming from genes [2–4]. The advantage of these methods lies in its huge inherent parallelism which has drawn great attention from the scientific community so far. In recent years, many different models of P systems have been proposed, such as cell-like P systems, tissue-like P systems, and spiking neural P systems [5–9]. The obtained computing systems prove to be so powerful that they are equivalent with Turing machines [10] even when using restricted combinations of features and also computationally efficient. Up to now a number of applications were reported in several areas such as biology, biomedicine, linguistics, computer graphics, economics, approximate optimization, cryptography, and so forth.
Traditionally, Morse theory is the subject of differential topology and differential geometry where the topological spaces in question are smooth manifolds. When we want to study discrete problems, we will use combinatorial complexes rather than manifolds. Along this line, discrete Morse theory has been developed [11, 12]. Recently, discrete Morse theory has attracted many researchers because it has found applications in triangulations and graphics. In fact, simplicial complex, the basic data structure in discrete Morse theory, will prove to be an important data structure besides trees and graphs.
Spatial cluster analysis is a traditional problem in knowledge discovery from databases [13]. It has wide applications since increasingly large amounts of data obtained from satellite images, X-ray crystallography, or other automatic equipment are stored in spatial databases. The most classical spatial clustering technique is due to Han and Kamber [13] who developed a variant PAM algorithm called CLARANS, while new techniques are proposed continuously in the literature aiming to reduce the time complexity or to fit for more complicated cluster shapes. Other clustering-like problems include impulsive cluster anticonsensus of discrete multiagent linear dynamic systems [14] and driving general complex networks into prescribed cluster synchronization patterns by using pinning control [15]. In another research, the authors introduce a cooperative article bee colony algorithm for solving clustering problems [16]. Also the authors propose a DNA-based clustering method by the Adleman-Lipton model [17]. For related research, one can also refer to [18–20].
In medical analysis there often appear data clustering problems of various type. The Wisconsin Breast Cancer Showhouse was founded in 1998 as an all-volunteer 501(c)(3) charitable organization by Nance Kinney, a breast cancer survivor (http://www.breastcancershowhouse.org/wbcs2012/index.html). Its mission is to support breast cancer and prostate cancer research at the Medical College of Wisconsin. This breast cancer databases were obtained from the University of Wisconsin Hospitals, Madison from Dr. William H. Wolberg. Many computing methods have been applied to study this data case and, in fact, the Wisconsin Breast Cancer data set is becoming an important testing benchmark for soft computing.
Inspired by the above research, this paper focuses on the joint study of discrete Morse theory with membrane computing. Our purpose is to propose a P system on simplicial complex. Up to our knowledge, this is the first paper to extend membrane computing to complexes. Then we use membrane computing in cluster analysis, providing a new approach to data mining. We first propose a discrete Morse structure for a candidate of a class of new P systems. Then we described for the first time a communication P system on simplices. Then we propose a new method for cluster analysis by simplicial P systems. Finally, we present the Wisconsin Breast Cancer analysis.
2. Discrete Morse Structure
In this section we present some general discrete models which will form the basis of membrane structures. The main idea comes from [11, 21]. In order to do this, we need to present some basic topological concepts. For simplicity we always assume that we are working in an Euclidean space Rn.
2.1. Simplex without Orientation
A k-simplex (cell) σ is the convex hull of k + 1 affinely independent points. More precisely, suppose a0, a1, …, ak are affinely independent, that is, a1 − a0, …, ak − a0 are linearly independent. Then σ is defined as the set of points in the form , where and λ0, λ1, …, λk ≥ 0. We will call k the dimension of a simplex and write dim (σ) = k, while a0, a1, …, ak are called vertices of the simplex. A simplex is uniquely indicated by its vertices and hence is expressed as σ = [a0, a1, …, ak], or simply σ = a0a1 ⋯ ak, and will be called a cell in this paper.
A face τ of a simplex σ is defined as a simplex generated by a nonempty subset of its vertices. We write τ < σ. A face τ is called a hyperface of σ if dim τ = dim σ − 1 and is denoted by τ≺σ. In this case, σ is called the parent of τ. Two cells τ1 and τ2 are called incident if τ1, τ2≺σ, and σ is called the coface of τ1 and τ2. Two cells σ1, σ2 are called neighbors if they share a common hyperface. The cone from a vertex x to a k-simplex σ is the convex hull of x and σ which yields a (k + 1)-simplex xσ provided x is not an affine combination of the vertices of σ. A simplicial complex K is a finite collection of nonempty simplices for which σ ∈ K and τ≺σ implies τ ∈ K and σ1, σ2 ∈ K implies that σ1∩σ2 is either empty or a face of both. The underlying space of K is the union of simplices: |K | = ⋃σ∈Kσ. Kp is a subset of K containing simplices of dimension p.
For a k-simplex σ, define K to be the collection of σ and all its faces. Then it is clear that K is a simplicial complex. We will call this simplicial complex a simple complex or, simply, a complex.
Now we consider some properties of incident and neighborhood relations as described above. First suppose τ1, τ2≺σ are incident and dim σ ≥ 2. Then τ1, τ2, σ are faces of a simplex. Define ψ = τ1∩τ2 and we will show that ψ ≠ ∅. This is evident because if , then p ≥ 2. By removing one vertex we obtain τ1 and τ2 with p vertices remained. Since p ≥ 2 we know that there exists at least one common vertex among τ1, τ2. By the definition of K we get ψ ∈ K and consequently τ1, τ2 are neighbors.
Conversely, if τ1, τ2≻ψ are neighbors, they need not be incident as shown in Figure 1. In the case when they are in the same simplex, however, this is true. In fact, suppose . Then all the vertices belong to τ1 and τ2. Then we can assume that and . Since both cells are in the same simplex, define . Then σ ∈ K. Consequently τ1, τ2 are incident. Putting everything together we get a theorem.
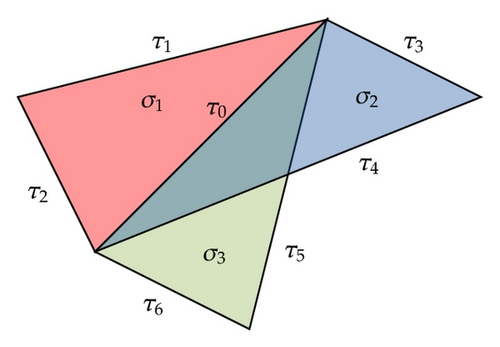
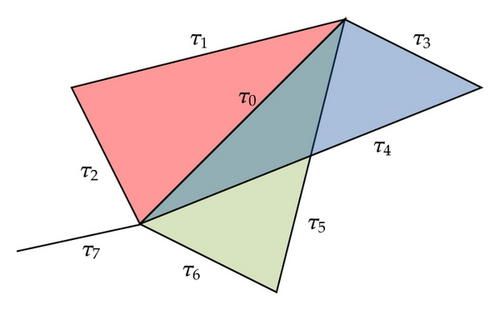
Theorem 2.1. Two incident cells τ1, τ2 are neighbors if they are at least one-dimensional. Two neighboring cells are incident provided they are located in the same simplex.
By the definition of neighborhood, if σ1, σ2 are neighbors and their common hyperface is τ, then clearly τ = σ1∩σ2 and, consequently, this common hyperface is unique. However, there exist cells with nonempty intersection but they are not neighbors. Now we consider incident cells τ1, τ2≺σ. We will show that their coface is also unique. First if dim τ1 = dim τ2 = 0, then dim σ = 1. Therefore σ is the edge joining the two vertices τ1, τ2 and hence is unique. Next if dim τ1 = dim τ2 ≥ 1, then as described previously we define ψ = τ1∩τ2. Then τ1 = v1ψ, τ2 = v2ψ and v1 ≠ v2. Therefore σ = v1v2ψ and is unique.
Theorem 2.2. Neighboring cells can only share a unique common hyperface. Incident cells have a unique coface (parent).
2.2. Simplex with Orientation
Simplicial complexes with orientation are important tools in the study of topological properties of discrete data structure. Concepts about discrete Morse functions are listed as follows (Robin [11]). For a simplex σ = [a0, a1, …, ak], there are two orientations and the opposite orientation is denoted by −σ. Figure 2 shows the orientation of a three-dimensional complex.
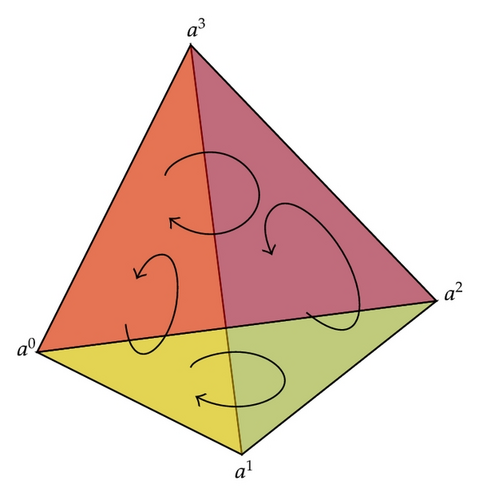
In simplicial complex we can define Morse functions which is a tool for optimization.
Definition 2.3. Let K be a simplicial complex. A function f : K → R is a discrete Morse function if for every σp ∈ Kp the following two statements are true:
-
(1) #{τp+1 > σ∣f(τ) ≤ f(σ)} ≤ 1; #{vp−1 < σ∣f(v) ≥ f(σ)} ≤ 1.
A simplex σp is critical with index p if
-
(2) #{τp+1 > σ∣f(τ) ≤ f(σ)} = 0; #{vp−1 < σ∣f(v) ≥ f(σ)} = 0.
A natural gradient flow is Φ(σ) = σ + ∂V(σ) + V(∂σ).
3. Communication P Systems on Simplices
3.1. Traditional P Systems
- (i)
m, m′ are vicinal, if m′ ⊂ m and there is no m′′ such that m′ ⊂ m′′ ⊂ m,
- (ii)
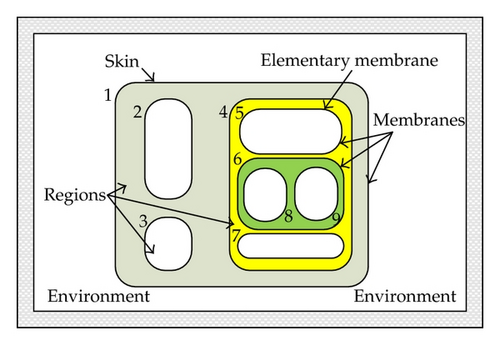
elementary membrane: with no lower vicinal membranes, skin membrane: with no upper vicinal membranes, we assume there is always a unique skin,
- (iii)
degree: number of membranes,
- (iv)
sibling membranes m, m′: if there is a m′′ which is upper vicinal for both m and m′.
A P system is called stable if, even if some rules are still applicable, their application does not change the string/object content of the membrane structure, nor the membrane structure itself. For a subalphabet W ⊂ O, we call a system stable over W if the projection over W of the string/object remains unchanged, even if some rules are still applicable. If W = {a}, we will say stable over a. If , with for 1 ≤ i ≤ m, is a subset of rules, we call a P system stable with respect to the rules R′ if the P system with rules R′ is stable (i.e., applications of rules in R′ do not change the string/object content of the system’s membranes) [19].
3.2. Membrane Structures on Simplices
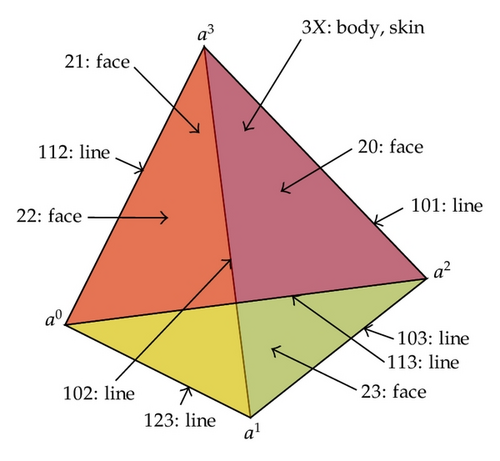
The boundary relations of simplices are shown in Figure 5 where the arrows point to the boundary cells.
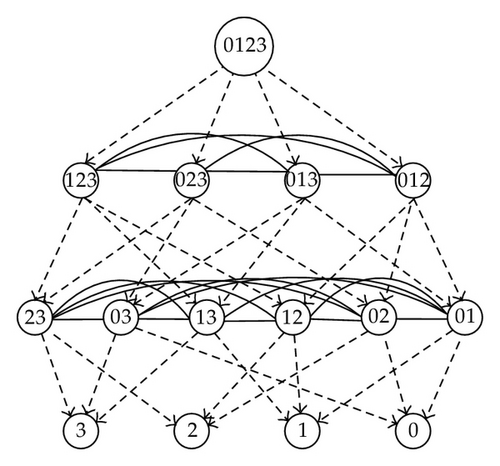

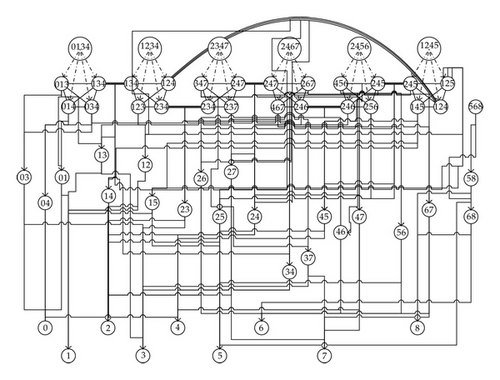
Generally speaking, a simplicial complex K is denoted by a set of vertices {a0, a1, …, ak} in Rn. A simplex σ ∈ K is called membrane. A membrane is called a max-simplex if it is not a face of another simplex in K. A simplex is denoted by its vertices. Evidently, vertices are zero-dimensional cells and hence are elementary membranes. If τ≺σ is a face of σ, then we way that σ is the parent of τ. Figure 5 shows the parents and neighborhood relations of the complex in Figure 4. For the more general simplicial complex as in Figure 6, the network model is shown in Figure 7.
If σ1, σ2 are incident, we say there is an upper link (channel) between σ1, σ2. If σ1, σ2 are neighbors, we say there is a lower link between them. A upper link is denoted by (σ1, σ2) τ, where τ is their (common) parent, while lower link is written as (σ1, σ2) τ, where τ is their common hyperface. Upper link is also written by (σ1, σ2) U or, simply, (σ1, σ2), while lower link is denoted by (σ1, σ2) L. Links have no directions. Thus (σ1, σ2) and (σ2, σ1) are identical. We will specify one from these two links, and only one is allowed.
Definition 3.1. A P system on a simplicial complex K, called a simplicial P system, with antiport and symport rules is a construct
Ceterchi and Martin-Vide [19] proposed a new type of communication P systems with priority relations. They introduced a promoter for a rule to be active and a inhibitor for it to be inactive. Induced by their idea, we will present an ordered system in this section.
An antiport rule (u, out; v, in) i in Ri exchanges the multiset u inside Ri with v outside it. A symport rule (x, out) i (or (x, in) i) sends out (takes in) the multiset x with respect to membrane Ri. For a specific membrane Ri, rules are totally ordered as r1 > r2 > ⋯>rn > rn+1 > ⋯. The rule rn+1 is applicable if and only if the system has reached a stable configuration with respect to rules r1 > ⋯>rn. We can use a queue structure to represent this process.
3.3. Rules in Simplicial P Systems
Now we describe the communication rules in simplicial P systems. For our purpose in this paper, there are mainly four types of communication rules in a simplicial P system. Each type of rule may have operators such as out, in, up, down.
First suppose (τ1, τ2) σ is an upper link. A rule like [(x, y), up] U means that the multiset x and y from τ1 and τ2 transform into z and go up to their parent σ. For two cells τ≺σ, the antiport rule [u, out; v, in∣τ≺σ] in τ means exchanging multiset u inside membrane τ with the multiset v outside it (in σ). The symport rule [u, out∣τ≺σ] sends the multiset u outside τ. Another symport rule [u, in∣τ≺σ] works similarly.
Equation (3.6) means moving x from cell τ1 and y from τ2 to z to parent σ and simultaneously moving γ from σ to α into τ1 and β into τ2. Equation (3.8) means moving x from cell τ1 and y from τ2 to z to hyperface σ and simultaneously moving γ from σ to α up to τ1 and β up to τ2.
3.4. Configuration and Computation
Now we describe the configuration and computation of simplicial P systems. For our purpose, change of membrane structure is not involved. A configuration of a simplicial P system is the state of the system described by specifying the objects and rules associated to each membrane. The initial state is called initial configuration. Therefore, the multisets represented by ωi, 1 ≤ i ≤ d in Π, constitute the initial configuration of the system.
The system evolves by applying rules in membranes and this evolution is called computation. The computation starts with the multisets specified by ω1, …, ωm in the m cells. In each time unit, rules are used in a cell. If no rule is applicable for a cell, then no object changes in it. The system is synchronously evolving for all cells.
When the system has reached a configuration in which no rule is any longer applicable, we say that the computation halts. A configuration is stable if, even if some rules are still applicable, their application does not change the object content of the membranes. The computation is successful if and only if it halts, or it is stable. The result of a halting/stable computation is the number described by the multiplicity of objects present in the cell i0 in the halting/stable configuration.
4. Cluster Analysis by Simplicial P Systems
4.1. Problem Setting and Algorithm
The clustering is implemented by a hierarchical method as follows. At first, each point of the data set X forms a cluster which contains a singleton. Then each data point tries to connect another data point in its neighborhood. After this step, a cluster can be found as connected points. Now we construct a simplicial complex K corresponding to the data set Ω. On the basis of the rectangles as in (4.1), we add some hyperplanes to form a triangulation. Then each rectangle is decomposed as several simplices. Hyperplanes can be chosen such that the set of simplices satisfy the definition of simplicial complex. And then K is defined as the union of such simplices. In the three dimensional case, this is shown as in Figure 8.
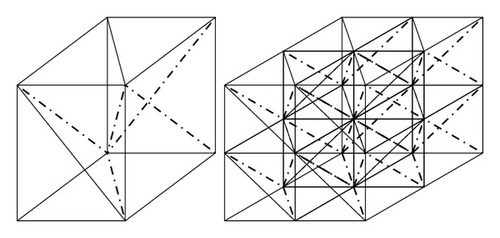
Then #(Sj) = 2n−1. Clearly C can be decomposed as disjoint cones . Notice that Sj is an (n − 1)-dimensional rectangle and this means that, for any triangulation of Sj, we can join them with v0 to form a triangulation of v0C. Therefore by induction we get the construction of triangulation.
Lemma 4.1. The decomposition is valid with the property that each pair of Int (v0Sj) is disjoint, where Int (C) is the set of interior points in Rn.
Proof. Suppose x = [a1, …, an] ∈ C and x ≠ v0. Consider the point xt = (1 − t)v0 + tx = [(1 − t)z1+ta1, …, (1 − t)zn + tan] where t ≥ 1. Let t0 be the first zero point of the following function:
Clearly ϕ(1) ≥ 0. Let . Then x ∈ v0SJ.
By the above discussion, we also have another lemma.
Lemma 4.2. Suppose the triangulation of Sj in Rn−1 is . Then the set forms a triangulation of C.
Finally we obtain a theorem.
Theorem 4.3. A simplicial complex K exists corresponding to the grids 𝒢.
For each node x ∈ Ω, define its neighborhood as 𝒩(x) = {y ∈ Ω∣y is incident with x in K}. For a subset C ⊂ Ω, define its neighborhood 𝒩(C) as 𝒩(C) = ⋃x∈C𝒩(x).
Now we propose an algorithm for our clustering problem. This is a self-joining cluster technique. Initially, one data point in Ω forms a cluster of singleton. Then each node searches for its neighborhood. If there exists a neighboring node which is similar enough, then activate a link between the two nodes. The final cluster is linked nodes. To define the meaning of similar enough, we need a parameter δ0 such that f(x, y) > δ0. Putting everything together, we get the algorithm as shown in Algorithm 1.
-
Algorithm 1: A self-joining clustering algorithm.
-
Inputs: Ω = {x1, x2, …, xN}, δ0: similarity threshold value, 𝒩i = {y∣y, xi are incident}
-
Outputs: 𝒞 = {C1, C2, …, Ct}: set of clusters, t: number of clusters, Ω0: outliers
-
Begin
-
Set Ω1 = Ω0 = ∅, C1 = ∅, t = 1.
-
for i = 1 to N
-
If 𝒩i = ∅, then add xi into set Ω0. Otherwise add xi into set Ω1. For each xj ∈ 𝒩i, j > i, calculate
-
the similarity measure f(xi, y). If f(xi, y) > δ0, then set the edge σ(xiy) as active. In this case, set
-
C1 = {xi} if C1 is still an empty set.
-
end
-
while Ω1 ≠ ∅ do
-
(1) For each x ∈ Ω1 if there exists an active edge σ(xy) with x ≠ y, y ∈ Ct, then add x into Ct and
-
remove x from Ω1.
-
(2) t = t + 1.
-
end
-
End
4.2. Design of a P System
Let vi = xi, i = 1, …, N be the vertices. We will use to denote a k-dimensional simplex. Therefore σ(vi) is a vertex or vi for short. Define Ni = #(𝒩i). ch is the set of edges in K.
5. Example and Discussion
The Wisconsin breast cancer data set consists of 699 samples with 9 attributes. The samples are classified as two categories, the malignant and the benign. The nine attributes are Clump-Thickness, Cell-Size-Uniformity, Cell-Shape-Uniformity, Marginal-Adhesion, Single-Epi-Cell-Size, Bare-Nuclei, Bland-Chromatin, Normal-Nucleoli, and Mitoses. The data is shown in Figure 9 with an added noise of [0,0.5].
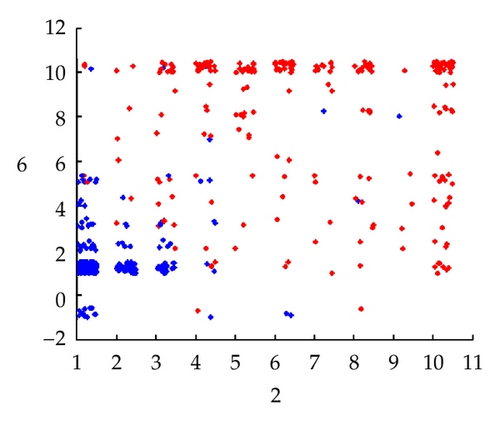
Next we construct a simplicial complex K as shown in Figure 10 in R2 where each vertex corresponds to the data points as in (5.1).
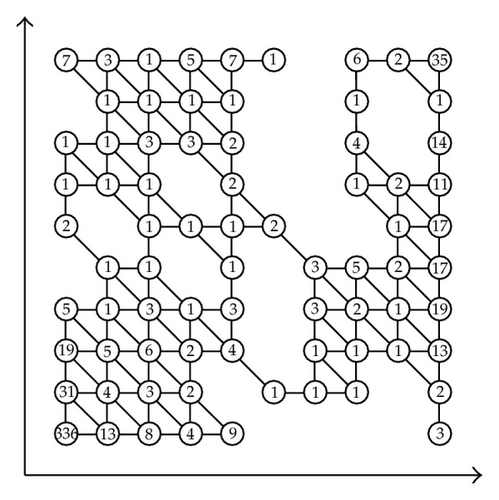
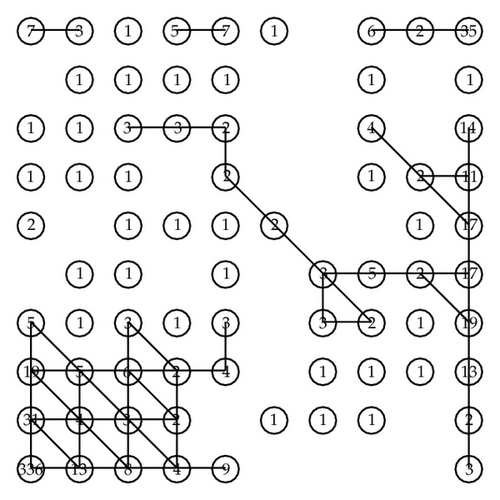
We now define L = 9. Then 2L = 512 and N = 100 − 25 = 75. Now we choose δ0 = 1. Then the final cluster is shown in Figure 11. We find five clusters and 31 outliers.
Now we analyze the effect of the parameter δ0. First we choose δ0 = 0. Then the clustering result is one cluster with all data. Then the ill clustered points is 699. Next let δ0 = 1. Then we get two clusters and some outliers. However, in this case the red data are all ill-clustered and hence the error rate is 241. Now choose δ0 = 2. In this case we find four clusters. The number of outliers are 46 while the ill-clustered points are 65 + 74 = 139. Then the total number is 185. Next choose δ0 = 3. This time we find 6 clusters. Number of outliers is 87. Number of ill-clustered points is 91. Number of other error points is 40 hence the total number is 218. Again we choose δ0 = 4. In this time we obtain 4 clusters with a large outlier (outlier number: 157, error clusters: 65, other errors: 13, total number: 235). The error line is shown in Figure 12.
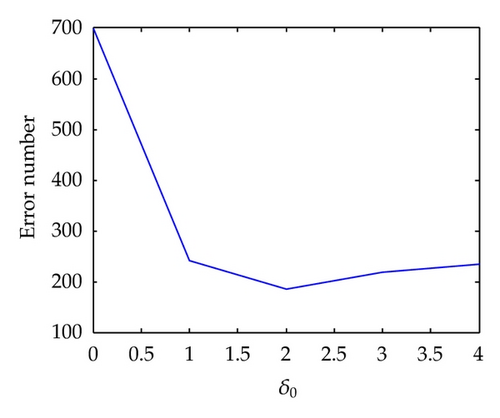
As a result we see that the best parameter is δ0 = 2.
Acknowledgments
This research is supported by the Natural Science Foundation of China (no. 61170038,71071090,60873058), the Natural Science Foundation of Shandong Province (no. ZR2011FM001), and the Shandong Soft Science Major Project (No. 2010RKMA2005).




