Evolutionary Selection of Features for Neural Sleep/Wake Discrimination
Abstract
In biomedical signal analysis, artificial neural networks are often used for pattern classification because of their capability for nonlinear class separation and the possibility to efficiently implement them on a microcontroller. Typically, the network topology is designed by hand, and a gradient-based search algorithm is used to find a set of suitable parameters for the given classification task. In many cases, however, the choice of the network architecture is a critical and difficult task. For example, hand-designed networks often require more computational resources than necessary because they rely on input features that provide no information or are redundant. In the case of mobile applications, where computational resources and energy are limited, this is especially detrimental. Neuroevolutionary methods which allow for the automatic synthesis of network topology and parameters offer a solution to these problems. In this paper, we use analog genetic encoding (AGE) for the evolutionary synthesis of a neural classifier for a mobile sleep/wake discrimination system. The comparison with a hand-designed classifier trained with back propagation shows that the evolved neural classifiers display similar performance to the hand-designed networks, but using a greatly reduced set of inputs, thus reducing computation time and improving the energy efficiency of the mobile system.
1. Introduction
The traditional way to craft an artificial neural network (ANN) for a classification task is to hand design a network topology and to find a set of network parameters using a gradient-based error-minimization algorithm such as back propagation [1]. However, in real-world applications, such as the classification of biomedical signals, the network topology can be difficult to design by hand. Additionally, in many cases, it is desirable to minimize the computational cost of the network, for example, by reducing the number of inputs used by the classifier. Evolutionary methods for the design of ANNs can provide an answer to both issues [2]. In this paper, we study the application of a neuroevolutionary method called analog genetic encoding (AGE) [3] to the problem of synthesis and optimization of neural networks for the processing of biological signals aimed at sleep and wake classification.
Continuous monitoring of the sleep/wake state of high-risk professionals such as pilots, truck drivers, or shift workers can potentially decrease the risk of accidents and help scheduling breaks and resting times. However, implementing such a classification in a wearable device is a challenging task. Limited energy and processing resources as well as the increased noise level due to movement artifacts and a constantly changing environment put tight restrictions on the choice of sensors and algorithms. Traditionally, the states of sleep and wake are classified based on the analysis of brain wave patterns (EEG) [4]. EEG recording requires gluing electrodes to the scalp and is typically susceptible to different sources of noise. Methods relying on EEG measurements are thus more suited for sleep analysis in controlled hospital environments than for mobile applications.
For mobile sleep/wake pattern screening, a commonly used technique is actigraphy [5]. In actigraphy, the acceleration of the wrist of the subject is recorded, and phases of weak activity—as judged by the levels of acceleration—are classified as sleep. Actigraphy devices can be small, inexpensive, and low power, which makes them suitable for mobile applications. However, as the signals provided by actigraphy devices are not directly linked to physiological states, it is difficult to derive a reliable prediction from them. Activities characterized by low levels of motion, such as reading or watching TV, are often misclassified as sleep [6]. In [7, 8], we have suggested to use electrocardiogram (ECG) and respiratory effort (RSP) signals for wearable sleep/wake classification (see Figure 1). Both signals depend on properties of the activity of the autonomous nervous system, which differ in sleep and wake [9]. Furthermore, they are measurable with portable sensor systems such as the Heally system (see Figure 2, Koralewski Industrie Elektronik, Celle, Germany).
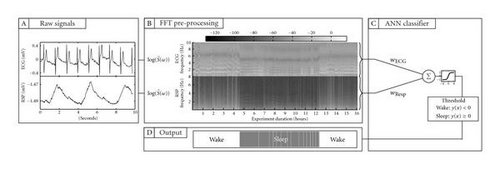

An additional difficulty is that the generation of a set of labeled data for the training of the classifier is typically a time-consuming activity for both the subjects from whom data is collected and the technicians who must label the data [7]. It is thus desirable to design a classifier that can be trained on a set of data and can then be used on further subjects without additional training. In [7], we have shown that using the frequency content of the ECG and RSP signals as input features for a single layer ANN, a mean accuracy of 86.7% can be achieved when the network was trained and tested on data obtained from different subjects. A limitation of the hand-designed ANN used in [7] is its large number of inputs. Some of these inputs are presumably redundant and might not contribute significantly to the classifier performance. For the targeted mobile application, the power consumption of the classifier is critical. In order to reduce processing time and thus power consumption for mobile applications, it would be desirable to minimize the number of inputs. In this paper, we show how to automatically synthesize networks that use a small subset of the spectral components associated with the signals as inputs while maintaining the performance of the classifier.
2. Evolutionary Synthesis of Neural Networks
Neural networks can be described as directed graphs, where the nodes represent a neuron model, and the edges of the graph are associated with the weighted connections between the neurons, the so-called synaptic weights. The design of a network for a particular task thus involves the choice of the topology of the graph (i.e., the network architecture) and a suitable set of numerical parameters (i.e., the synaptic weights and the parameters of the neuron model). The automatic synthesis of the topology and parameters of a neural network requires a computer representation for both aspects of the network, combined with an algorithm capable of performing a search in the space defined by this representation. Evolutionary algorithms have been extensively used to evolve neural classifiers because these algorithms can combine a flexible representation with a high potential of stochastic exploration of the search space [10–13].
The simplest approach to this, the so-called direct encoding, represents all the neurons, synaptic connections, and parameters of the network explicitly (see, e.g. [14–16]). This has the advantage that the resulting networks can easily be decoded from the genome. However, with increasing size of the network, the length of the corresponding genome grows rapidly, which can affect the evolvability. In order to mitigate this problem, it has been suggested to encode a program or a sequence of instructions that, when executed, builds the network. This developmental encoding can lead to very compact representations of large networks (see, e.g., [17, 18]). However, the definition of a set of mutation and recombination operators which guarantees that only valid networks are generated during the search is typically very difficult.
A promising alternative to direct and developmental representations that is getting more and more popular is implicit encoding [19–23]. In this paper, we use an implicit representation called analog genetic encoding (AGE). AGE has been shown to be very effective for the automatic synthesis of various kinds of networks and, in particular, of neural networks [2, 3, 24–26].
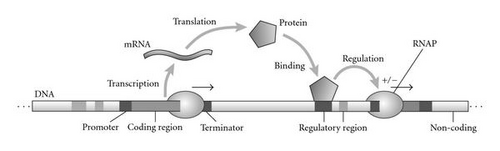
The concept of implicit encodings like AGE is loosely inspired by the working of biological gene regulatory networks (GRNs). In biological GRNs, the interactions between the genes are not explicitly encoded in the genome but follow implicitly from the physical and chemical environment in which the genome is immersed. Simplifying a bit the picture, the activation of a biological gene depends on the interaction of molecules produced by another gene with parts of the activated gene called regulatory regions (Figure 3(a)). AGE abstracts this picture and defines an artificial genome composed of sequences of characters, for example, the uppercase ASCII set (Figure 3(b)). Similar to the function of promoter and terminal regions in GRNs, special sequences (the so-called tokens) identify regions of the artificial genome as artificial genes, which encode individual neurons. The sequences delimited by the tokens are interpreted analogous to coding regions and regulatory regions in biological GRNs. The strength of the connection between two neurons is implicitly determined by the coding region of one neuron and the regulatory region of another neuron via a function called interaction map. The interaction map can be seen as an abstraction of the biochemical process of gene regulation. It takes sequences of characters as arguments and outputs a real-valued connection strength. In our implementation, this is obtained by mapping the local alignment score [27] of the two sequences exponentially to the interval that spans all possible weight values (see [24]).
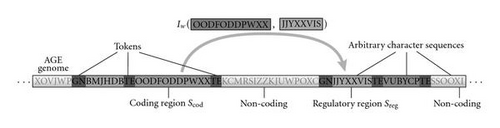
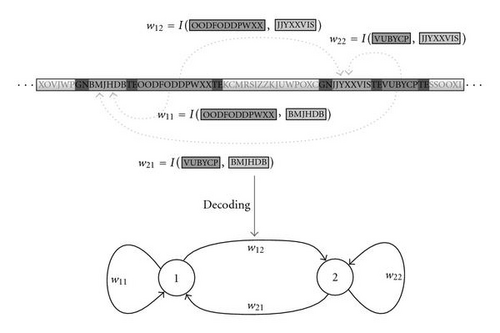
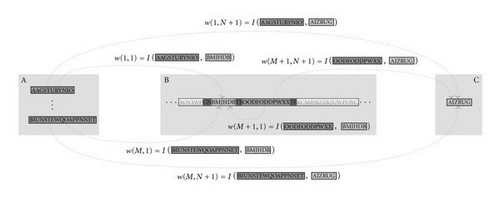
In summary, the AGE genome can be decoded first by extracting the neurons with the associated (coding and regulatory) sequences of characters. This is realized by scanning the genome for tokens which indicate the presence of a neuron (GN). Together with predefined terminator sequences (TE), these tokens delimit the part of the genome associated with the respective neuron. The enclosed sequences of characters are interpreted as the coding and regulatory sequences of the respective neuron. Subsequently, the interaction map I can be applied to all pairs of coding and regulatory sequences to obtain the synaptic weights wij connecting the neurons (see Figure 4).
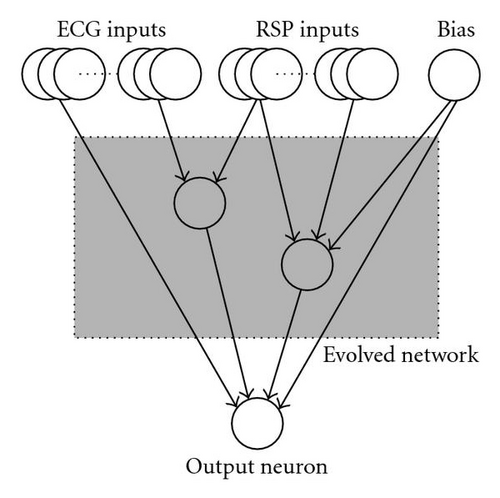
In this framework, there are several different possibilities to implement connections from external inputs to external outputs (see [28] for more details). Here, we encoded the coding sequences associated to the input neurons and the regulatory sequences associated to the output neurons in separated parts of the genome (see Figure 5). In this case, the connections from the input neurons to the network can be obtained by applying the interaction map to all pairs of coding sequences (associated with the input neurons and the hidden neurons) and regulatory regions (associated to the hidden neurons and the output neuron). Note that the interaction map can associate a null weight value, thus leaving the respective neurons unconnected. When this feature is applied to the connections stemming from the input neurons, it gives evolution the freedom to select a subset of the set of inputs that contains the information necessary to realize the classification task.
As the sequences which define the strength of the synaptic connections can have a variable length and the interaction map is defined to operate on sequences of arbitrary length, a large class of genetic operators can be used to alter the network. In particular, we use the biologically plausible insertion, substitution, and deletion of characters and the transposition, duplication, and deletion of fragments of genome. The changes in the genome caused by these mutation operators can reflect both changes in the parameters of the network as well as changes in the network structure. For example, the insertion of a character in the genome can lead to a change of the synaptic weight connecting a particular input to the output neuron. The deletion of a fragment of genome associated with an input of the network can lead to the removal of this particular input from the network. Furthermore, the number of hidden neurons in the network can increase (e.g., after a genome fragment duplication) or decrease (e.g., after a character substitution) over the course of evolution. Given the fact that parts of the genome can be noncoding (i.e., they are not part of the description of a neuron) and that the interaction map is defined to be highly redundant, many mutations do not have an effect on the decoded networks. This allows for a high neutrality in the search space, which can improve evolvability [29].
3. Experiments
To compare the performance of the classical approach to classifier synthesis and training with the state-of-the-art neuroevolution method based on AGE, we performed a set of experiments, where we compared the performance of a neural network with fixed hand-designed topology and variable weights trained with back propagation, with that of neural networks synthesized with an evolutionary algorithm-based on AGE. As anticipated, we are interested in the performance in a sleep/wake detection task, where data from a set of users is available for network synthesis and training, but the performance is expected to generalize to additional users. We thus investigated the performance of the two methods when trained on ECG and RSP data collected on multiple subjects, and tested on data from a different subject.
3.1. Data
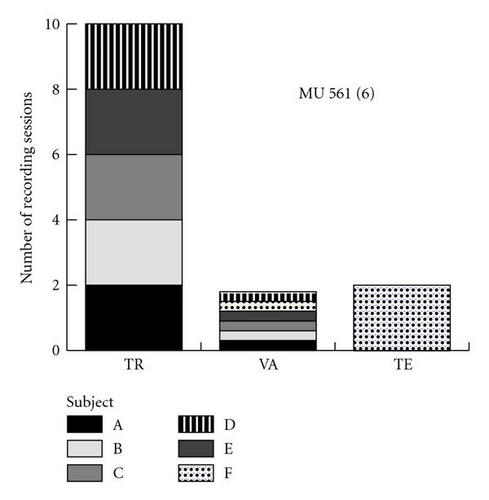
The data used in the following experiments are identical with those described in [8], where a hand-designed classifier with back propagation was used. They stem from recording sessions with six young healthy male subjects of a mean(± SD) age of 26(± 3) years. The subjects wore a Heally recording device (see Figure 2) for a total of 18 recording sessions which lasted 16 hours each and contained an overnight sleep. The datasets are composed of ECG and RSP recordings sampled at 100 Hz and 50 Hz, respectively. The a priori sleep and wake states of the subjects were determined by a trained technician who labeled the signals in 10-second intervals based on electromyogram, electrooculogram, and video recordings. The data were preprocessed and fed to the ANN. As in [7], the preprocessing step consisted of calculating the power spectrum of each signal using a short-time fast Fourier transform with a window length of 40.96 seconds (see Figure 1(b)). For each of these segments, we calculated a feature vector as where is the periodogram of the segment. Experiments described in [8] revealed that frequency components above 10 Hz for ECG and 8 Hz for RSP do not contribute to the hand-designed classifier performance and can be removed. The resulting two input vectors are thus composed of 409 spectral inputs from ECG and 327 spectral inputs from RSP. Together, they compose the set of 736 inputs that were fed to the ANN classifier (see Figure 1(c)).
3.2. Experimental Design
In order to evaluate the performance of the two classifiers, we divided the data into three different sets: atraining set (TR), a validation set (VA), and a test set (TE) (see Figure 6). The training set contains a subset of the data from five of the six subjects. The validation set is composed of 2 hours of data from each subject, randomly sampled over the two available sessions and containing an equal amount of samples labeled as sleep and wake. This data is not used for training or for testing. The test set contains data from the subject that has not been used in the training. Five independent runs of each experiment are performed from different randomly assigned initial conditions. In order to prevent performance biases due to the choice of sessions, we repeat each experiment with all possible combinations of users in the test and trainingsets, making sure that the same sessions do not appear both in the training and in the testsets. This leads to a total of six different cases with five independent replications for each case.
3.3. Algorithms
3.3.1. Hand-Designed Fixed Topology Network
As a baseline for the classification accuracy, we used a feed-forward ANN with no hidden layers and a single output unit with a tangent-sigmoid transfer function. Additional experiments not reported here showed that the use of ANNs with a hidden layer does not improve the performance of the classifier. A similar finding has been reported by [30]. The synaptic weights of this fixed topology network were initialized with the Nguyen-Widrow method [31] and trained with a Levenberg-Marquardt back-propagation algorithm [32].
3.3.2. Network Synthesized with Age
Selection was performed using tournament selection and elitism. The algorithm parameters and mutation probabilities are listed in Table 1. In order to prevent bootstrap problems, the population was initialized with the best 100 networks out of 1000 randomly created genomes. Additionally, to save computation time, only a randomly selected subset of 10% of each training set was used for training. However, validation and testingwere always performed using 100% of the respective dataset. For each evolutionary run, the synthesized network was the network with the best performance on the validation set, in the collection of all the best performing networks observed at each of the 1000 generations that compose a run.
| Parameter | Value |
|---|---|
| Population size | 100 |
| Tournament size | 2 |
| Elite size | 1 |
| Recombination probability | .1 |
| Probability of character substitution (per character) | .001 |
| Probability of character insertion (per character) | .001 |
| Probability of character deletion (per character) | .0015 |
| Probability of fragment transposition | .01 |
| Probability of fragment duplication | .01 |
| Probability of fragment deletion | .015 |
| Probability of neuron insertion | .01 |
For both the back-propagation training and the evolutionary process, the measure of quality of the classifier was the sum over the data points of the squares of the difference between the actual and the desired classifier output.
4. Results and Discussion
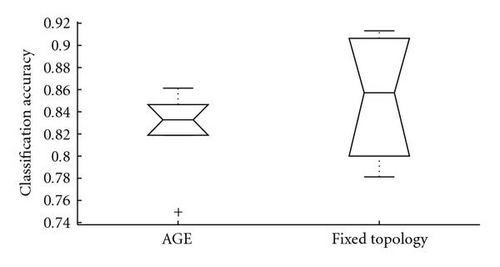
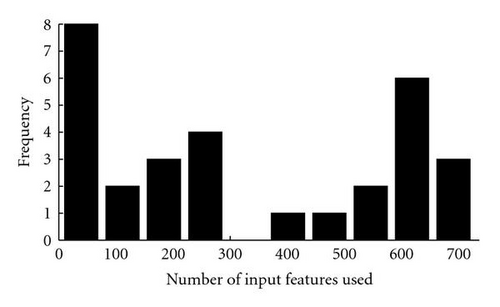
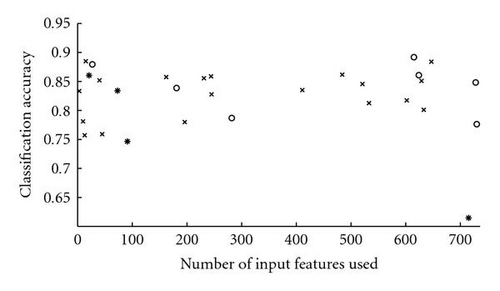
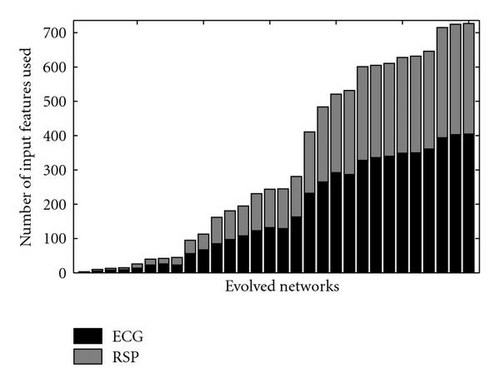
As shown in Figure 8, the evolved networks and the fixed topology networks trained with back propagation do not display a significantly different classification accuracy (Wilcoxon rank sum test P = .48). However, while the hand-designed fixed topology networksemploy all of the 736 input features, many of the evolved networks used a drastically reduced set of inputs (see Figure 9, the median of the number of inputs used is 244.5). Figure 10 shows that there is no correlation between the number of inputs used by the evolved networks and their performance (Spearman′s rank correlation coefficient P = .02, P = .94). This indicates that many input features are indeed redundant and that it is possible to synthesize networks with a very small number of inputs which perform as well as the hand-designed network using all inputs. However, all networks use input features from both ECG and RSP data (see Figure 11). Given the results of [7], it is not surprising that the presence of both types of data is beneficial for the classification accuracy and thus selected during evolution. Note that in the evolutionary experiments, no additional penalty term was added to the objective function to bias the search toward small networks. This explains the presence of both networks using a significantly reduced set of inputs, and networks using almost the whole set of available inputs in the evolutionary results.
As mentioned above, the fixed topology network has no hidden layer. Of the 30 evolved networks, 19 feature no hidden neurons, 7 feature one hidden neuron, and 4 feature two hidden neurons. However, there is no correlation between the number of hidden neurons and the classification accuracy (Spearman′s rank correlation coefficient P = −.06, P = .74). This substantiates the conjecture formulated in [7] that a hidden layer is not necessary for optimal performance in this task. Note, however, that this conjecture applies to this specific problem and does not extend to general classification applications.
5. Conclusion
Portable devices for biomedical signal analysis, like sleep/wake classification, have the potential to alleviate health problems and prevent accidents. Recent advances in sensor development and miniaturization allow for the construction of small mobile devices which integrate biomedical sensors and a microprocessor with sufficient processing power for many applications. However, one of the critical challenges, that remains, is the design of efficient classifiers which can be implemented on these small mobile systems. While the classification accuracy has to be as high as possible, the computational effort and thus the energy requirements for classification have to remain low. The results presented in this paper demonstrate that analog genetic encoding (AGE) permits the automatic evolutionary synthesis of compact neural classifiers for the problem of sleep/wake classification. Compared to a hand-designed classifier trained with back propagation, the possibility of the evolutionary selection of a subset of the available inputs permits a drastic reduction of the number of inputs without significant degradation of the classifier performance. For example, in the experiments presented here, the evolutionary synthesis with AGE found a classifier with the accuracy of 88.49%, using only 15 of the 736 input features used by the hand-designed network. The implementation of this evolved solution on a digital signal controller of the dsPIC33 product family (Microchip Technology Inc., USA) requires only 5.13% of the instructions used by an implementation of the hand-designed network on the same processor. This is a reduction of the computational cost of almost 95%. Moreover, the savings in computational cost and energy can be increased even further by adapting the sensory modalities and preprocessing steps to the reduced set of input features.
Acknowledgments
This work was supported by the Swiss National Science Foundation, Grant no. 200021-112060 and the Solar Impulse Project grant of Ecole Polytechnique Fédérale de Lausanne (EPFL). Thanks to Daniel Marbach and Sara Mitri for their comments on an earlier version of this manuscript and the anonymous reviewers for their helpful suggestions.




