Circulation of vaccinia virus in southern and south-eastern wildlife, Brazil
Abstract
We evaluated 345 wild animals from southern and south-eastern Brazil to understand their role in vaccinia virus (VACV) transmission cycle. VACV DNA was detected in rodents, marsupials, chiroptera and cingulate, expanding the knowledge of VACV host range in wildlife that could potentially act as source of infection in rural and urban areas.
1 INTRODUCTION
Vaccinia virus (VACV) is a zoonotic agent of wide geographic distribution in several countries of South America, especially in Brazil where it is responsible for a disease called bovine vaccinia (BV) (Oliveira et al., 2017). BV has been predominantly reported in rural areas, affecting mainly dairy cattle and humans, being a burden to public health and local dairy economies (Oliveira et al., 2017). Additionally, VACV has also been detected in other species present in the rural environment (equids and buffaloes) (Lima et al., 2019) and domestic animals in urban areas (dogs and cats) (Costa et al., 2018; Oliveira et al., 2017). However, VACV natural history and circulation in wildlife are still poorly explored (Miranda et al., 2017; Oliveira et al., 2017).
Few studies have documented the circulation of VACV in synanthropic and wild rodents (Abrahão et al., 2009; Miranda et al., 2017), and also in other sylvatic animals such as primates, marsupials and coatis (Abrahão et al., 2009; Costa et al., 2018; Lima et al., 2019; Miranda et al., 2017; Oliveira et al., 2017). Aiming to include the wildlife in the VACV transmission cycle, ecological models have been proposed (Costa et al., 2017, 2018; Lima et al., 2019; Oliveira et al., 2017). According to the models, sylvatic animals could potentially act in VACV transmission between rural, wild and even urban environments (Abrahão et al., 2009; Costa et al., 2018; Lima et al., 2019; Oliveira et al., 2017). Furthermore, VACV could also circulate silently in areas with very few cattle herd and anthropogenic disturbance (Kurth et al., 2008). Hence, studies focused on VACV circulation in wildlife could add information regarding its ecoepidemiology, assessing the risks of VACV spreading from the wild to rural and urban areas, as well documented for Cowpox virus in Europe (Essbauer, Pfeffer, & Meyer, 2010; Kroon et al., 2016).
2 MATERIALS AND METHODS
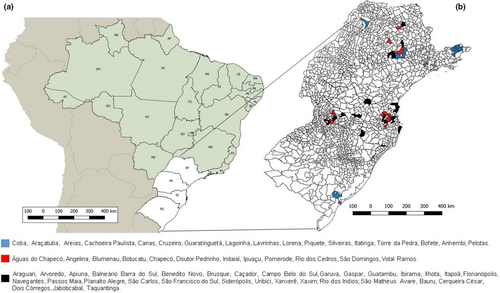
We retrospectively analysed a total of 345 DNA samples extracted from the liver of wild animals captured during 2007–2011 in southern and south-eastern areas of Brazil (Figure 1). In these sampled areas, VACV outbreaks are frequently reported in São Paulo state and Rio Grande do Sul state. This study was approved by the Animal Ethics Committee of the Instituto Adolfo Lutz (protocol no. 2/2015 and 2/2017) and Animal Ethics Committee of the Faculdade de Medicina Veterinária e Zootecnia (protocol no. 211/2008). Moreover, it is in accordance with the Brazilian Institute of Environment and Renewable Natural Resources’ (IBAMA) normative statement n. 119 of 11 October 2006, chapter VI, art 0.26. Liver samples were manually fractionated with disposable surgical blades or scalpels and processed mechanically by using Mini-Beadbeater-24 BioVortexerTM homogenizer (Biospec Products, Bartlesville, USA). Samples were submitted for DNA extraction using the Illustra Tissue & Cells genomic Prep Mini Spin kit (GE Healthcare, Chicago, USA) in Instituto Adolfo Lutz and send to Laboratório de Vírus at Universidade Federal de Minas Gerais (UFMG). To detect VACV DNA, we performed a semi-nested PCR targeting the C11R gene (viral growth factor; vgf) and a real-time PCR to A56R gene (hemagglutinin) has been used in genetic analyses for VACV differentiation (Kroon et al., 2016; Peres et al., 2018).
We directly sequenced the amplified fragments in both orientations and in duplicate by using the ABI3130 platform (Applied Biosystems—Thermo Fisher Scientific, Foster City, USA). Sequences were aligned with other reference sequences from GenBank by using MEGA 7.0 software.
3 RESULTS AND DISCUSSION
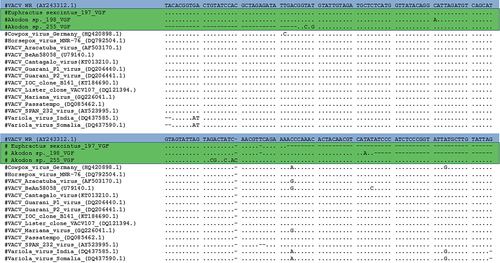
A large diversity of wild animals such as rodents, non-human primates, marsupials, carnivores, chiropterans, placental mammals, lagomorph, cingulate and artiodactyl mammals were screened (Table 1). A total of 12 animals (3.5%) being rodents, non-human primates, marsupials and cingulate tested positive for C11R gene, in which Euphractus sexcintus and Akodon sp. had amplicons sequenced. Alignment of the C11R fragments showed highly similarity (90%–100%) to the homologous gene of orthopoxviruses, including VACV strains isolated from Brazil (Figure 2). Additionally, 18 animals (5.2%) tested positive for A56R gene. Of these, we detected the VACV in two bats (Molossus rufus and Eumops perotis species), both sampled in Botucatu city, São Paulo state. The VACV circulation in bats has never been explored in Brazil before. Alignment of the amplified A56R fragments showed 100% similarity to the homologous gene of VACV isolates from Brazil (Figure 3) and the presence of an 18-nt signature deletion, which is present in sequences of mouse non-virulent Brazilian-VACV strains (group 1 Brazilian VACV).
| Order | Families | Species | No. of tested animals | No. of positive animals | |
|---|---|---|---|---|---|
| C11R | A56R | ||||
| Artiodactyla | Cervidae | 1 | 0 | 0 | |
| Carnivora |
Felidae Canidae Mustelidae Procyonidae |
Cerdocyon thous, Procyon cancrivorus, Nasua nasua, Galictis cuja Leopardus trigrinus, Puma yagouaroundi Leopardus wiedii, Puma concolor Lontra longicaudis |
23 | 0 | 0 |
| Chiroptera |
Molossidae Phyllostomidae |
Eumops perotisƗ,Tadarida brasiliensis Molossus molossus, Molossus rufusƗ Eumops glaucinus, Artibeus obscurus Artibeus lituratus |
20 | 0 | 2 |
| Cingulata | Dasypodidae |
Euphractus sexcintusƗ Dasypus novemcinctus |
3 | 1 | 0 |
| Didelphimorphia |
Didelphidae Marmosidae |
Didelphis albiventris, Monodelphis sp. Micoureus paraguayanus Philander frenatus, Didelphis aurita Gracilinanus microtarsusƗ Lutreolina crassicaudataƗ |
31 | 2 | 3 |
| Lagomorpha |
Leporidae |
Lepus europaeusƗ | 5 | 0 | 1 |
| Pilosa |
Myrmecophagidae |
Tamandua tetradactyla Myrmecophaga tridactyla |
12 | 0 | 0 |
| Primates |
Cebidae Atelidae Callitrichidae Cercopithecidae |
Saimiri sciureusƗ, Alouatta guariba Callithrix sp., Alouatta belzebul Alouatta seniculus, Ateles chamek Ateles marginatus, Ateles paniscus Cebus albifrons, Erythrocebus patas Lagothrix lagotricha, Alouatta caraya Mandrillus sphinx, Alouatta fusca Papio hamandryasƗ, Papio papio |
47 | 1 | 1 |
| Rodentia |
Echimyidae Muridae Cricetidae Caviidae Sciuridae Erethizontidae |
Nectomys sp., Akodon montensisƗ Hydrochoerus hydrochaerisƗ Oligoryzomys nigripes, Cavia aperea Thaptomys nigritaƗ, Delomys sp., Oxymycterus sp., Scapteromys sp. Brucepattersonius iheringi Euryoryzomys russatusƗ Sooretamys angouya Guerlinguetus aestuansƗ Sooretamys angouya Nectomys squamipesƗ Sphiggurus spinosusƗ, Rattus rattusƗ |
203 | 8 | 11 |
Note
- Ɨ: Positive species in molecular screening
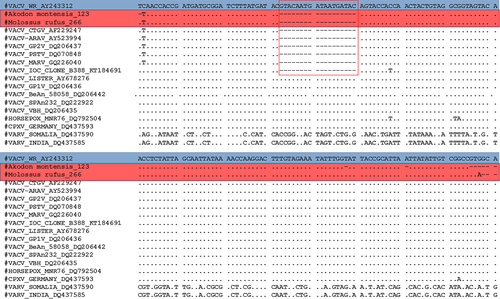
These findings reinforce the hypothesis that VACV can widely circulate in wild environments in Brazil, potentially infecting a wide range of hosts. The detection of VACV in Akodon montensis, a species with generalist habits, corroborates the participation of rodents in VACV epidemiological cycle as previously described (Abrahão et al., 2009; Miranda et al., 2017). Furthermore, the detection of VACV in Molossus rufus, a forest bat that also possesses synanthropic habits, reinforces the hypothesis that these wild animals could act as sources of transmission of wildlife viruses (Abrahão et al., 2009; Costa et al., 2018). It is important to emphasize that the wildlife animals included in this study were sampled in areas with ecosystem alterations due to anthropic actions, such as agricultural activity and hydroelectric dam plant construction. These factors could enhance the occurrence of generalist and/or peridomestic species, whose VACV circulation has already been described (Miranda et al., 2017), as well as bringing other wild animals closer to rural and urban areas, thereby expanding their potential to act as sources of VACV exposure to humans and domestic animals. A similar dynamic is well established about the natural circulation of Cowpox virus (CPXV) in Europe. The dynamic comprise wild rodents identified as natural hosts and felines act as intermediate hosts enabling the transmission of CPXV to other animals and even humans (Essbauer et al., 2010; Kurth et al., 2008).
Limitations of our study include lack of clinical information from the sampled animals aiming to evaluate the presence of clinical signs suggestive of VACV infection, absence of serum samples that could allow us estimate the seroprevalence in the wildlife and the absence of viable clinical material for virus isolation. However, our findings provide important information regarding the circulation of VACV in wild environments, contributing to understand the VACV maintenance in the wildlife in the absence of outbreaks. Additional studies should investigate the potential of transmission of specific groups of wild animals (other than rodents) to fully elucidate their role in VACV epidemiological cycle.
ACKNOWLEDGEMENTS
We thank colleagues from the Laboratório de Vírus (ICB-UFMG) for their excellent technical support. Financial support was provided by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) and Pró-Reitoria de Pesquisa/UFMG (PRPq-UFMG). EG Kroon, JS Abrahão and GS Trindade are researchers from CNPq.
CONFLICT OF INTEREST
None to declare.