RETRACTED: The causal relationship between immune cells and atopic dermatitis: A bidirectional Mendelian randomization study
Abstract
Background
Atopic dermatitis (AD) is a chronic inflammatory skin condition whose origins remain unclear. Existing epidemiological evidence suggests that inflammation and immune factors play pivotal roles in the onset and progression of AD. However, previous research on the connection between immune inflammation and AD has yielded inconclusive results.
Methods
To evaluate the causal relationship between immunological characteristics and AD, this study employed a bidirectional, two-sample Mendelian randomization (MR) approach. We utilized large-scale, publicly available genome-wide association studies to investigate the causal associations between 731 immunological feature cells and the risk of AD.
Results
Significant associations were identified between six immune phenotypes and AD risk: increased Basophil %CD33dim HLA DR−CD66b−, CD25 on IgD+ CD24+, CD40 on monocytes, HLA DR on CD14+ CD16−monocytes, HLA DR on CD14+monocytes correlated with higher AD risk, while elevated CD3 on CD4 Treg was linked to lower risk. Reverse MR analysis revealed AD as a risk factor for IgD+ CD38br AC and IgD+ CD38br %B cell, but a protective factor against CD20 on IgD+ CD38− naive and CD8 on NKT.
Conclusion
Our findings elucidate the intricate interplay between immune cells and AD, informing future research into AD pathophysiology and therapeutics.
1 INTRODUCTION
Atopic dermatitis (AD) is a chronic inflammatory skin disease primarily affecting the flexural surfaces of the limbs, with localized or widespread manifestations, resulting in varying degrees of decreased quality of life for patients.1, 2 Most patients are prone to allergies and often have concurrent allergic rhinitis, allergic asthma, allergic conjunctivitis, and other atopic diseases.3, 4 AD is an autoimmune disorder clinically characterized by dry, itchy skin, polymorphic erythema, and papules, with a tendency for exudation during severe flare-ups. Laboratory parameters such as serum total IgE and eosinophil count are significantly associated with AD exacerbations. While most cases of AD manifest before school age, approximately 25% of patients experience clinical symptoms in adulthood, with slight variations in clinical presentation across different age groups.5 AD has a high global incidence, affecting around 25% of children and 10% of adults, with a rising trend in recent years due to its environmental associations, leading to variable incidence rates in different regions.6 Currently, systemic treatments for AD mainly include corticosteroids, immunosuppressants, biologics, and JAK inhibitors. AD exerts a substantial negative impact on patient's physical and mental health and imposes significant economic burdens.7 Hence, early diagnosis and intervention can mitigate disease severity, thus improving the prognosis of AD.
The pathogenesis of AD is complex and diverse, and not yet fully understood. Immune cells have been shown to be associated with a variety of skin diseases. A Mendelian randomization (MR) analysis showed that CD62L on dendritic cells and HLA-DR on monocytes are risk factors for alopecia areata, which may provide new options for the treatment of alopecia areata.8 Similarly, another MR analysis also demonstrated a correlation between lymphocytes and psoriasis.9 Existing extensive research indicates a crucial role of immune factors in the etiology of AD. The balance between immune cells Th1 and Th2 forms the foundation for maintaining normal immune function, whereas AD involves Th2 cell over-differentiation, regulating the expression of Th2-mediated cytokines and chemokines, leading to Th1/Th2 immune imbalance and compromised skin barrier integrity.10, 11 IL-4 and IL-13 are two major Th2 cell cytokines associated with the pathogenesis of AD. Following skin damage, mast cells induce IL-13, which directly inhibits IL-12, a Th1-related interleukin, and is subsequently released by CD4+ T cells, mediating the immune response against antigens.12, 13 Additionally, the JAK/STAT signaling pathway holds significance in the pathogenesis of AD and inflammation.14 The JAK family consists of four intracellular kinases: JAK1, JAK2, JAK3, and TYK2, while the STAT family involved in signal transduction comprises seven members: STAT1, STAT2, STAT3, STAT4, STAT5a, STAT5b, and STAT6.15 The IL-4/IL-13-dependent JAK/STAT pathway involves STAT6 and STAT3 proteins, upregulating the expression of T2 specific transcription factor GATA3 and promoting B cell class switching to IgE, thereby regulating T-cell proliferation and T2 differentiation.16
With advancements in high-throughput sequencing technology, our understanding of the immune system's function and the key cell types involved in AD pathogenesis continues to grow. IL-18 promotes T2-mediated inflammation by activating eosinophils and mast cells. Elevated IL-18 levels correlate with increased AD incidence, while IL-18 deficiency in mouse models improves AD skin lesions by reducing IL-4 levels and mast cell infiltration.17, 18 Additionally, innate and adaptive immune cells, such as eosinophils, basophils, and macrophages, play crucial roles in AD pathogenesis. Eosinophils contribute to AD initiation through increased IL-4 expression and interactions with keratinocytes and dermal macrophages, resulting in epidermal hyperplasia and impaired skin barrier function.19 Circulating eosinophils increase in AD patients, along with elevated IL-4 and IL-13 production mediated by the JAK/STAT pathway.20 Studies indicate upregulation of IL-18 receptors in eosinophils of AD patients, with histamine enhancing expression of H2R and H4R in eosinophils, suggesting critical roles of IL-18 and histamine in eosinophil-mediated inflammation.21 In the cutaneous lesions of AD, there is notable infiltration of immune cells expressing IL-31, such as M2 macrophages, CD68+ macrophages, and eosinophils. The number of IL-31-expressing M2 macrophages correlates positively with TSLP and periostin expression in AD lesion epidermis and disease severity. Depletion of eosinophils in AD mouse models leads to decreased IL-31 expression in M2 macrophages.22 Furthermore, M2 macrophages produce C-C motif chemokine ligand 18 (CCL18), a chemokine closely associated with increased AD incidence.23 However, current research on the causal relationship between AD and immune inflammation yields inconsistent results, possibly due to small sample sizes, confounding factors, and biases in study designs. Presently, topical corticosteroids are the most common treatment for AD, used by 60% of patients.24 Despite their benefits in alleviating AD symptoms and reducing inflammation, long-term use may lead to resistance and side effects such as epidermal atrophy, growth retardation, and impaired wound healing.25 Although biologics and small molecule drugs for AD exist, they do not fully control symptoms and progression. Hence, further research into inflammatory targets in AD is necessary to identify more effective treatment options and improve patients' quality of life.
In general, the primary criterion for establishing causality is randomized controlled trials (RCTs). However, conducting an RCT is inherently complex, requiring significant participant involvement and resources. At times, ethical constraints may render investigating a particular factor nearly impossible. MR studies present an effective alternative approach.26 Genome-wide association studies (GWAS) employ high-throughput genomic techniques to identify variations associated with traits or diseases in specific populations, including single nucleotide polymorphisms (SNP) and copy number variations (CNV), thereby enhancing our understanding of the complex genetic characteristics of various diseases.27 Two-sample Mendelian randomization (2SMR) is a research method using summary data from GWAS to estimate the causal impact of exposure on outcomes. In 2SMR analysis, SNP, also known as instrumental variables(IVs), are employed to analyze the causal relationship between exposure and outcomes. From a genetic standpoint, SNP is randomly allocated from parents to offspring, akin to randomization in controlled trials conceptually.28 Furthermore, 2SMR studies can mitigate the effects of reverse causality, and eliminate confounding factors' interference, rendering the studies more reliable and trustworthy. The association between certain immune cells and AD has been identified in earlier investigations.29, 30 In this study, we conducted an extensive two-way, two-sample MR analysis to elucidate the causal relationship between immune cells and AD.
2 MATERIALS AND METHODS
2.1 Study design
This study employed bidirectional two-sample MR to investigate causal associations between 731 immune cell phenotypes and AD risk, using large-scale publicly available GWAS data. The reliability of the MR results in this study relies on three critical assumptions: (1) SNP closely correlates with immune cells; (2) SNP is unrelated to known confounding factors; (3) SNP affects AD solely through immune cells and is not directly associated with AD.31, 32
2.2 Data sources
The exposure and outcome data utilized in this study were obtained from publicly available databases. The exposure factor consisted of 731 immune cells, with data retrieved from the public repository (GCST0001391 to GCST0002121), encompassing GWAS data for the 731 immune phenotypes included in the study.33 Additionally, data for AD (ID: ebi-a-GCST90018784) were sourced from the IEU Open GWAS project database, comprising 481,299 cases and 475,075 controls of European descent, with 24,185,642 SNP.34 All data involved in this study were derived from public databases, thus negating ethical review concerns.
Previous studies have employed a significance threshold of p < 1 × 10−5 to select significant SNPs for various immune traits.35, 36 Leveraging the European 1000 Genomes Project, the CLUMP program in PLINK software was utilized to exclude IVs with r2 < 0.001. For reverse MR analysis, a significance threshold of 5 × 10−8 and r2 of 0.001 were set. An F statistic greater than 10 indicates the unlikely presence of weak instrumental variable bias.
2.3 Statistical analysis
This study utilized three MR methods: inverse variance weighted (IVW), MR-Egger regression, and weighted median analysis. Odds ratio (OR) and 95% confidence interval (CI) were employed as evaluation metrics, with a significance level set at α = 0.05. IVW was considered the primary analytical approach when all genetic variations met instrumental variable assumptions, providing an appropriate estimation of causal effects.30 However, IVW results may be influenced by certain pleiotropic biases, necessitating sensitivity analysis for result validation. In cases of heterogeneity detected in sensitivity testing, random-effects IVW was adopted. MR-Egger regression served as a robust assessment tool when the assumption of independence between the direct effects of IVs and outcome factors and the association effects between IVs and exposure factors held true.37 Weighted median analysis indicated validity when IVs contributed more than 50% to genetic variance.38 Among the statistical values obtained from these three two-sample MR methods, IVW was chosen as the main analytical outcome. Additionally, scatter plots demonstrated the minimal impact of outliers on results, while funnel plots depicted the robustness of correlations in the absence of heterogeneity.
3 RESULTS
3.1 Exploring the impact of immune phenotype on the causal relationship of AD
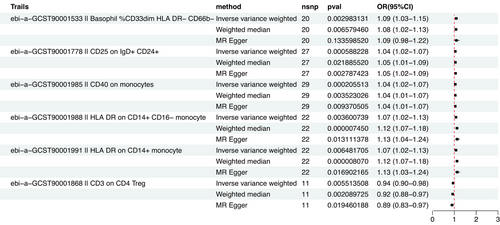
As the primary analytical approach, we employed two-sample MR analysis and the IVW method to investigate the causal relationship between immune phenotypes and AD. Data heterogeneity assessment was conducted using Cochran's Q test, with the fixed-effects model IVW applied when p > 0.05. Based on p < 0.05, we found 44 immunophenotypes that were causally related to AD, and we selected six immunophenotypes with high significance, namely, Basophil %CD33dim HLA DR− CD66b−, CD25 on IgD+ CD24+, CD40 on monocytes, HLA DR on CD14+ CD16− monocyte, HLA DR on CD14+ monocyte, and CD3 on CD4 Treg.
Among them, the OR of Basophil %CD33dim HLA DR−CD66b− against AD calculated by the IVW method was 1.09 (95% CI: 1.03∼1.15; p = 2.98 × 10-3). Results from other MR methods were similar: weighted median (OR, 1.08; 95% CI: 1.02∼1.13; p = 6.58 × 10−3), and MR-Egger (OR, 1.09; 95% CI: 0.98−1.22; p = 0.133). The OR of CD25 on IgD+ CD24+ against AD calculated by the IVW method was 1.04 (95% CI: 1.02 ∼ 1.07; p = 5.88 × 10-4). Results from other MR methods were similar: weighted median (OR, 1.05; 95%CI: 1.01∼1.09; p = 0.02), and MR-Egger (OR, 1.05; 95% CI: 1.02−1.09; p = 2.8 × 10-3). The OR of CD40 on monocytes against AD calculated by the IVW method was 1.04 (95% CI: 1.02∼ 1.07; p = 2 × 10-4). Results from other MR methods were similar: weighted median (OR, 1.04; 95% CI: 1.01∼1.07; p = 3.5 × 10−3), and MR-Egger (OR, 1.04; 95% CI: 1.01−1.07; p = 9.37 × 10-3). The OR of HLA DR on CD14+ CD16− monocyte against AD calculated by IVW method was 1.07 (95% CI: 1.02∼1.13; p = 3.6 × 10-3). Results from other MR methods were similar: weighted median (OR, 1.12; 95% CI: 1.07∼1.18; p = 7.45 × 10−6), and MR-Egger (OR, 1.13; 95% CI: 1.04−1.24; p = 0.013). The OR of HLA DR on CD14+ monocyte against AD calculated by the IVW method was 1.07 (95% CI: 1.02∼1.13; p = 6.48 × 10-3). Results from other MR methods were similar: weighted median (OR, 1.12; 95% CI: 1.07∼1.18; p = 8 × 10−6), and MR-Egger (OR, 1.13; 95% CI: 1.03−1.24; p = 0.017). The OR of CD3 on CD4 Treg against AD calculated the by IVW method was 0.94 (95% CI: 0.90 ∼ 0.98; p = 5.5 × 10-3). Results from other MR methods were similar: weighted median (OR, 0.92; 95% CI: 0.88 ∼ 0.97; p = 2 × 10−3), and MR-Egger (OR, 0.89; 95% CI: 0.83−0.97; p = 0.019) (Table 1).
| Exposure | Outcome | Method | nSNP | p | OR (95% CI) |
|---|---|---|---|---|---|
|
Basophil %CD33dim HLA DR− CD66b− |
Atopic dermatitis |
Inverse variance weighted Weighted median MR-Egger |
20 20 20 |
2.98E−03 6.58E−03 0.133 |
1.09 (1.03–1.15) 1.08 (1.02–1.13) 1.09 (0.98–1.22) |
|
CD25 on IgD+ CD24+ |
Atopic dermatitis |
Inverse variance weighted Weighted median MR-Egger |
27 27 27 |
5.88E−04 0.02 2.8E−03 |
1.04 (1.02–1.07) 1.05 (1.01–1.09) 1.05 (1.02–1.09) |
| CD40 on monocytes | Atopic dermatitis |
Inverse variance weighted Weighted median MR-Egger |
29 29 29 |
2E−04 3.5E−03 9.37E−03 |
1.04 (1.02–1.07) 1.04 (1.01–1.07) 1.04 (1.01–1.07) |
| HLA DR on CD14+ CD16− monocyte | Atopic dermatitis |
Inverse variance weighted Weighted median MR-Egger |
22 22 22 |
3.6E−03 7.45E−06 0.013 |
1.07 (1.02–1.13) 1.12 (1.07–1.18) 1.13 (1.04–1.24) |
| HLA DR on CD14+ monocyte | Atopic dermatitis |
Inverse variance weighted Weighted median MR-Egger |
22 22 22 |
6.48E−03 8E−06 0.017 |
1.07 (1.02–1.13) 1.12 (1.07–1.18) 1.13 (1.03–1.24) |
| CD3 on CD4 Treg | Atopic dermatitis |
Inverse variance weighted Weighted median MR-Egger |
11 11 11 |
5.5E−03 2E−03 0.019 |
0.94 (0.90–0.98) 0.92 (0.88–0.97) 0.89 (0.83–0.97) |
- Abbreviation: OR, odds ratio.
While some MR-Egger tests yielded p > 0.05, the Beta values for the three aforementioned analytical methods were all > 0, indicating they all aligned in the same direction. The main distinction between the MR-Egger method and IVW lies in the consideration of the intercept term in the regression. This term is used to gauge the average magnitude of pleiotropy among IVs, while the slope provides an unbiased estimate of the causal effect. Generally, the standard error of the IVW method is smaller than that of the MR-Egger method.
Therefore, as the gold standard for MR analysis, IVW results will be prioritized in the absence of heterogeneity and horizontal pleiotropy. In summary, MR analysis remains statistically significant. There is no evidence of horizontal pleiotropy in the analysis of the present study. Sensitivity analysis further confirmed the robustness of causal associations derived from the analysis (Table 2, Figure 1).
| Exposure | Outcome | Method | Q | Q_df | Q_pval |
|---|---|---|---|---|---|
| Basophil %CD33dim HLA DR− CD66b− | Atopic dermatitis |
Inverse variance weighted MR-Egger |
48.08 48.03 |
19 18 |
0.00025 0.00015 |
| CD25 on IgD+ CD24+ | Atopic dermatitis |
Inverse variance weighted MR-Egger |
24.16 23.08 |
26 25 |
0.57 0.57 |
| CD40 on monocytes | Atopic dermatitis |
Inverse variance weighted MR-Egger |
38.74 38.73 |
28 27 |
0.09 0.07 |
| HLA DR on CD14+ CD16− monocyte | Atopic dermatitis |
Inverse variance weighted MR-Egger |
38.74 38.73 |
28 27 |
0.09 0.07 |
| HLA DR on CD14+ monocyte | Atopic dermatitis |
Inverse variance weighted MR-Egger |
42.28 38.83 |
21 20 |
0.004 0.007 |
| CD3 on CD4 Treg | Atopic dermatitis |
Inverse variance weighted MR-Egger |
8.28 6.16 |
10 9 |
0.56 0.72 |
3.2 Exploring the impact of AD on the causal relationship of immune phenotype
Reverse MR analysis detected statistical significance (p < 0.05) between AD and 77 immune phenotypes. We selected four immunophenotypes with high significance. Specifically, AD was identified as a risk factor for two immune phenotypes: IgD+ CD38br AC and IgD+ CD38br %B cell, while AD was found to be a protective factor for two immune phenotypes, namely CD20 on IgD+ CD38− naive and CD8 on NKT.
Among them, the OR of AD against IgD+ CD38br AC calculated by the IVW method was 1.05 (95% CI: 1.00 ∼ 1.10; p = 0.032). Results from other MR methods were similar: weighted median (OR, 1.07; 95%CI: 0.99∼1.17; p = 0.095), and MR-Egger (OR, 1.08; 95% CI: 1.01∼1.16; p = 0.034). The risk ratio of AD to IgD+ CD38br %B cell was estimated to be 1.05 (95% CI: 1.00∼1.10; p = 0.03). The results of the other three methods were similar: weighted median (OR, 1.07; 95% CI: 0.99∼1.14; p = 0.084), and MR-Egger (OR, 1.09; 95% CI: 1.01−1.17; p = 0.024). The risk ratio of AD to CD20 on IgD+ CD38− naive was estimated to be 0.94 (95% CI: 0.88∼0.99; p = 0.033). The results of the other three methods were similar: weighted median (OR, 0.93; 95% CI: 0.82∼1.06; p = 0.287), and MR-Egger (OR, 0.89; 95% CI: 0.81−0.98; p = 0.018). The risk ratio of AD to CD8 on NKT was estimated to be 0.94 (95% CI: 0.90∼0.99; p = 0.014). The results of the other three methods were similar: weighted median (OR, 0.94; 95% CI: 0.86∼1.04; p = 0.223), and MR-Egger (OR, 0.93; 95% CI: 0.86−1.00; p = 0.048) (Table 3).
| Exposure | Outcome | Method | nSNP | p | OR (95% CI) |
|---|---|---|---|---|---|
|
IgD+ CD38br AC |
Atopic dermatitis |
Inverse variance weighted Weighted median MR-Egger |
154 154 154 |
0.032 0.095 0.034 |
1.05 (1.00–1.10) 1.07 (0.99–1.17) 1.08 (1.01–1.16) |
| IgD+ CD38br %B cell |
Atopic dermatitis |
Inverse variance weighted Weighted median MR-Egger |
154 154 154 |
0.03 0.084 0.024 |
1.05 (1.00–1.10) 1.07 (0.99–1.14) 1.09 (1.01–1.17) |
| CD20 on IgD+ CD38− naive | Atopic dermatitis |
Inverse variance weighted Weighted median MR-Egger |
152 152 152 |
0.033 0.287 0.018 |
0.94 (0.88–0.99) 0.93 (0.82–1.06) 0.89 (0.81–0.98) |
| CD8 on NKT | Atopic dermatitis |
Inverse variance weighted Weighted median MR-Egger |
152 152 152 |
0.014 0.223 0.048 |
0.94 (0.90–0.99) 0.94 (0.86–1.04) 0.93 (0.86–1.00) |
- Abbreviation: OR, odds ratio.
Although some MR-Egger and weighted median tests yielded p > 0.05, the Beta values for the three aforementioned analytical methods were consistent in direction, thus MR analysis retains statistical significance. There is no evidence of horizontal pleiotropy in the analysis of the present study. Sensitivity analysis further confirmed the robustness of the causal relationships derived from the analysis (Table 4 and Figure 2).
| Exposure | Outcome | Method | Q | Q_df | Q_pval |
|---|---|---|---|---|---|
| IgD+ CD38br AC | Atopic dermatitis |
Inverse variance weighted MR-Egger |
177.92 176.76 |
153 152 |
0.08 0.08 |
| IgD+ CD38br %B cell | Atopic dermatitis |
Inverse variance weighted MR-Egger |
173.66 172.06 |
153 152 |
0.12 0.13 |
| CD20 on IgD+ CD38− naive | Atopic dermatitis |
Inverse variance weighted MR-Egger |
150.14 148.27 |
151 150 |
0.50 0.52 |
| CD8 on NKT | Atopic dermatitis |
Inverse variance weighted MR-Egger |
147.61 147.29 |
151 150 |
0.56 0.55 |

4 DISCUSSION
Our large-scale bidirectional MR study clarified intricate causal relationships between multiple immune phenotypes and AD, substantiating the pivotal role of immune dysregulation in AD pathogenesis.
According to this study, as the proportion of Basophil %CD33dim HLA DR−CD66b−, CD25 on IgD+ CD24+, CD40 on monocytes, HLA DR on CD14+ CD16−monocyte and HLA DR on CD14+ monocyte, the risk of AD increases. The risk of AD decreased as the proportion of CD3 on CD4 Treg increased. Previous studies have shown that basophilic granulocyte infiltration can be observed in about 60% of patients with AD.39 Basophils are circulating immune cells that are partially implicated in itchiness, and capable of promoting allergic skin inflammation. In the context of AD models, basophils are systemically activated and contribute to immune dysregulation.19, 40, 41 And Basophil %CD33dim HLA DR−CD66b− may be a potential cell subtype associated with atopic dermatitis. Research findings indicate an association between AD and various B-cell subtypes in both adult and pediatric patients.42, 43 However, existing literature has not yet identified changes in the expression levels of CD25 on IgD+ CD24+ B cells in AD, warranting further investigation. Monocytes maintain vascular homeostasis. Research indicates that in patients with AD, monocytes exhibit abnormal expression of inflammatory cytokines and antibodies, potentially exacerbating cutaneous inflammation.44, 45 Genetic association analyses reveal a causal relationship between CD14+ CD16- monocyte immune cells and Juvenile idiopathic arthritis.46 Furthermore, relevant studies have demonstrated abnormal expression of CD40 on monocytes in patients with primary humoral immunodeficiency.47 In a study employing flow cytometry, notable differences were observed in the expression levels of TLR2 within CD14(+) HLA-DR(+) PB monocytes between patients with AD and healthy controls.48
The pathogenesis of AD suggests that infiltration of helper type 2 (Th2) cells can release interleukin (IL)-4 and IL-13, leading to cutaneous inflammatory responses, thus highlighting the correlation between T cells and the onset of AD.49, 50 A study revealed that children with severe AD have a significantly reduced percentage of CD4(+) T cells expressing IL-4 compared to healthy controls, indicating its protective role in AD.51
Furthermore, the reverse MR studies have identified associations between AD and four immune cells: IgD+ CD38br AC, IgD+ CD38br %B cell, CD20 on IgD+ CD38− naive, and CD8 on NKT. In this context, AD can lead to increases in IgD+ CD38br AC and IgD+ CD38br %B cell (or > 1), while causing decreases in CD20 on IgD+ CD38− naive and CD8 on NKT (or < 1).
Multiple studies have indicated a correlation between abnormal B cell function and the onset of AD, even proposing it as a therapeutic approach for treating AD.43, 52 A study using a mouse model of AD demonstrated that administering anti-IgD therapy shortly after allergen exposure significantly reduced skin inflammation, suggesting a certain correlation between IgD on B cells and the pathogenesis of AD.53 A study on four-color flow cytometry reveals significant alterations in the quantity of iNKT cells and the cytokines they produce in patients with AD.54
Our dual-sample MR study constitutes a statistically robust analysis with substantial sample size, leveraging existing large-scale GWAS cohorts encompassing over 481,299 individuals diagnosed with Alzheimer's disease (AD) alongside European populations exhibiting immunological characteristics. To ascertain causal relationships between genetic variants associated with environmental factors and outcomes, MR serves as an epidemiological research tool. Given that exogenous confounders do not influence genetic variation, measurement errors in genetic variants and their effects can be disregarded, rendering the study findings overall credible. However, our study is not without limitations. First, the applicability of this finding remains somewhat restricted as the GWAS data utilized in this study are limited to European populations. Second, to obtain more precise analytical results, we were unable to conduct additional population sub-stratification analysis on current AD cohorts due to the lack of individual patient information within the GWAS data. Last, our study employed bidirectional MR analysis. While this approach greatly aids in addressing issues of causality, the interpretation of MR results must also consider biological mechanisms; relying solely on statistical effect estimates is insufficient.55 If stronger biological evidence could be incorporated to support it, the substantive causal relationship between immune cells and AD would be more thoroughly investigated, as current investigations into immunological characteristics of AD patients are not yet sufficiently comprehensive.
Nonetheless, our work provides genetic insights into AD etiology and potential immunological therapeutic targets, guiding future translational research toward developing more effective treatments and improving patient outcomes.
5 CONCLUSION
In conclusion, we have further elucidated the interaction patterns between AD and the immune system and investigated causal relationships between several immune phenotypes and AD utilizing bidirectional MR analysis. Bidirectional MR analysis was employed in our study to mitigate reverse causality, thus enhancing the accuracy of causal inference and reducing the influence of additional confounding factors. Apart from providing valuable insights for AD treatment, this also offers compelling genetic data for the study of AD's pathological physiology and biological functionalities.
ACKNOWLEDGMENTS
The study received no funding.
CONFLICT OF INTEREST STATEMENT
There is no conflict of interest in the study.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.




