RETRACTED: Identifying the genetic association between common rheumatic diseases and vitiligo
Abstract
Background
Although observational studies have suggested a correlation between vitiligo and rheumatic diseases, conclusive evidence supporting a causal relationship is still lacking. Therefore, this study aims to explore the potential causal relationship between vitiligo and rheumatic diseases.
Methods
Using genome-wide association studies, we performed a two-sample Mendelian randomization (MR) analysis. In our analysis, the random-effects inverse variance weighted (IVW) method was predominantly employed, followed by several sensitivity analyses, which include heterogeneity, horizontal pleiotropy, outliers, and “leave-one-out” analyses.
Results
The genetically predicted vitiligo was associated with an increased risk of rheumatoid arthritis (RA) (OR, 1.47; 95% confidence interval [CI], 1.29–1.68; p < 0.001), and systemic lupus erythematosus (SLE) (OR, 1.22; 95% CI, 1.06–1.39; p = 0.005). The causal associations were supported by sensitivity analyses. In Sjögren's syndrome and ankylosing spondylitis, no causal relationship with vitiligo was found in the study.
Conclusion
Our MR results support the causal effect that vitiligo leads to a higher risk of RA and SLE. Individuals with vitiligo should be vigilant for the potential development of RA and SLE. Managing and addressing this potential requires regular monitoring.
1 INTRODUCTION
Vitiligo is a disease in which white, depigmented spots appear on the skin due to the selective loss of melanocytes.1 The prevalence of vitiligo varies between 0.5% and 2.0% worldwide, with variations among different geographic locations and ethnic groups.2 Although it does not have a negative impact on life expectancy, it can reduce the self-esteem of patients, generate a great deal of psychological distress, and thus negatively affect the quality of life of patients.3-5 The exact pathogenesis of vitiligo remains complicated and unclear. Research suggests that genetics, autoimmunity, and oxidative stress are involved,2, 6 with autoimmunity being considered to be the primary contributing factor.
Rheumatic diseases encompass a spectrum of chronic conditions affecting the skeletal system, joints, surrounding soft tissues, and other associated structures and organs. This category comprises over 100 distinct disorders classified into 10 categories. The etiology remains elusive, with many of these ailments closely linked to autoimmune reactions. Rheumatic diseases can manifest as either localized pathological damage or systemic disorders. Failure to promptly address these conditions poses a significant risk of disability or even mortality, imposing substantial burdens on both society and families.
Patients with vitiligo are often accompanied by a variety of autoimmune diseases, such as alopecia areata, psoriasis, SLE, RA, thyroid disease, inflammatory bowel disease, and type 1 diabetes. Recently, a Mendelian randomization (MR) analysis conducted by Chen et al. suggested a genetic causal association between vitiligo and AITDs.7 Observational studies have revealed that patients with vitiligo have a markedly elevated risk of developing rheumatic diseases.8-12 So far, it remains uncertain if vitiligo and rheumatic diseases are causally related. Since observational studies are unable to establish causality because of unmeasured and uncontrolled confounding factors, we applied a two-sample MR analysis to further explore the causal connection between vitiligo and common rheumatic diseases such as rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), Sjögren's syndrome (SS) and ankylosing spondylitis (AS).
2 METHODS
2.1 Study design
To identify a possible causative link between vitiligo and four rheumatic diseases—RA, SLE, SS, and AS—a two-sample MR analysis was employed. The underlying assumptions of the MR analysis are as follows: first, selected exposure exhibits a strong correlation with instrumental variables (IVs). Second, the IVs and potential confounding factors must be independent; finally, the IVs can only contribute to results through exposure factors.
2.2 Data sources
To conduct our MR study, we obtained the genetic IVs for each trait from available genome-wide association studies (GWAS), as shown in Table S1. For the outcome datebase, genetic IVs for RA and SLE were obtained from the IEU OpenGWAS database (https://gwas.mrcieu.ac.uk/). For RA, which includes 14,361 cases and 43,923 controls; for SLE, which includes 5201 cases and 9066 controls. Genetic IVs for SS (2495 cases and 3,65,533 controls), and AS (2860 cases and 270,964 controls) were extracted from the Finn Gen Biobank (https://www.finngen.fi/en). For the exposure datebase, we extracted genetic IVs for vitiligo from the largest GWAS meta-analyses, which include 4680 cases and 39,586 controls.13 The cases and controls were all of European ancestry in this study.
2.3 Genetic instrumental variable selection
First, we extracted SNPs (single-nucleotide polymorphism) with a significant threshold of p-value < 5×10−8 from the full GWAS data. To ensure independent SNPs (kb = 10,000, r2 < 0.001), IVs with linkage disequilibrium (LD) were removed by employing the clump_data method. After identifying SNPs related to the exposure of interest (vitiligo), we extracted SNP instruments from the outcome GWAS dataset (RA, SLE, SS, and AS), respectively. IVs were eliminated if they demonstrated a strong correlation with the outcome phenotype. After that, we harmonized the effect estimates for both exposure and outcome variants and excluded palindromic SNPs. Finally, to evaluate the strength of the IVs, we employed the F-statistics, and an F-statistic > 10 was considered necessary.
2.4 MR analyses
In the MR analysis, the random-effects inverse variance weighted (IVW) method was employed as the primary analysis method, which allowed for heterogeneity across SNPs.14 We also utilized two other different MR techniques (weighted median [WM] and MR-Egger) to evaluate the reliability of the findings. Additionally, to confirm the main analysis's robustness, we carried out several sensitivity analyses. First, to assess whether there was heterogeneity among IVs, Cochran's Q-tests were conducted using the IVW and MR-Egger approaches. Interestingly, when total heterogeneity was balanced, the existence of heterogeneity did not affect the random-effects IVW estimation results. Potential heterogeneity was indicated when p < 0.05. MR-Egger intercepts were employed to assess potential horizontal pleiotropy; a p-value > 0.05 indicates the absence of any horizontal pleiotropy.15 The MR-PRESSO method was employed to evaluate potential pleiotropy and correct possible outliers; a p-value > 0.05 indicates the absence of pleiotropy.16 To assess the potential influence of a sole outlier variant on the effect estimates, we carried out a leave-one-out analysis.
2.5 Statistical analysis
The“TwoSampleMR” software package (version 0.5.8) was used to conduct a two-sample MR analysis. and the MR-PRESSO test was carried out using the “MRPRESSO” software package (version 0.5.8). R software (version 4.3.2) was used to perform all statistical analyses. To infer genetic causation, a significance threshold of p < 0.05 was used. To be specific, when p < 0.05 and Odds Ratio (OR) > 1, it suggests the existence of a positive genetic causality, while OR < 1 indicates negative genetic causality.
3 RESULTS
3.1 MR analysis
The IVs used for MR analyses are presented in Table S2. As shown in Figure 1, a positive genetic causal connection between vitiligo and RA was found by the IVW model (OR = 1.47, 95% confidence interval [CI]: [1.29−1.68], p < 0.001). Moreover, similar effect estimates were obtained by the WM techniques. With the WM method, the causal relationship between vitiligo and RA (OR, 1.28; 95% CI: 1.09−1.50; p < 0.01) is demonstrated, which is consistent with the IVW method (Figure 1). We carried out the sensitivity analysis to evaluate the dependability of the results (Table S3). The results of the Cochran's Q test showed p-values less than 0.05, indicating that there was heterogeneity. The MR Egger intercept test revealed no horizontal pleiotropy (p > 0.05). In the MR-presso test, no outliers were identified. The results of the leave-one-out analysis demonstrate that no single outlier variant had an impact on the genetic assessment (Figure 2A). The above results suggest that, compared with non-vitiligo patients, vitiligo patients had a 1.47-fold increased risk of RA.
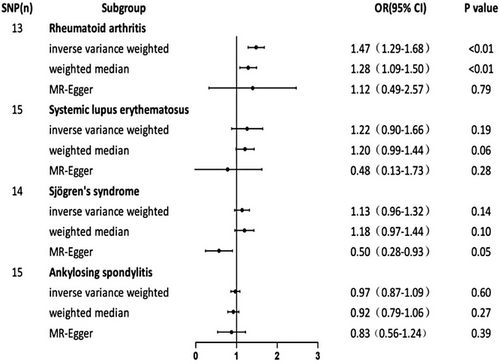

When SLE was employed as the outcome variable, the IVW method's combined OR estimate was 1.22 (95% CI: 0.90−1.66, p = 0.19) (Figure 1). Results of sensitivity analysis between SLE and vitiligo show significant heterogeneity by Cochran's Q statistic (p < 0.05). There was no horizontal pleiotropy found by the MR Egger intercept test (p > 0.05). However, the MR-presso test suggested the presence of horizontal pleiotropy (p < 0.05) and identified three outliers (Table S3). After eliminating the three potential outliers that were previously mentioned, we carried out the IVW analysis of random effects once more. Interestingly, there was significant genetic causation between vitiligo and SLE (p = 0.005, OR 95% CI: 1.22 [1.06–1.39] (Table 1). No heterogeneity and horizontal pleiotropy were shown by the sensitivity analysis (Table 2). The results of the leave-one-out analysis demonstrate that no single outlier variant had an impact on the genetic assessment (Figure 2B). These results suggest that, compared with non-vitiligo patients, people with vitiligo had a 1.22-fold higher risk of developing SLE.
| Method | SNPs | p-value | OR (95% CI) |
|---|---|---|---|
| Inverse variance weighted | 12 | 0.005 | 1.22(1.06–1.39) |
| Weighted median | 12 | 0.062 | 1.19(0.99–1.44) |
| MR-Egger | 12 | 0.141 | 1.69(0.89–3.20) |
- Abbreviations: 95% CI, the 95% confidence intervals; OR, odds ratio; SLE, systemic lupus erythematosus; SNP, single-nucleotide polymorphism; MR, Mendelian randomization.
| Test | Method | p-value |
|---|---|---|
| Heterogeneity | Cochran's Q test (IVW) | 0.57 |
| Cochran's Q test (MR-Egger) | 0.58 | |
| Pleiotropy | Intercept test (MR-Egger) | 0.33 |
- Abbreviations: IVW, inverse variance weighted; MR, Mendelian randomization; SLE, systemic lupus erythematosus.
When SS and AS were employed as outcome variables, the IVW method's combined OR estimate was 1.13 (95% CI: 0.96−1.32, p = 0.14) in SS, and 0.97 (95% CI: 0.87−1.09, p = 0.60) in AS, respectively (Figure 1). This result suggests that there is no significant genetic causal relationship between vitiligo and SS, AS. The outcomes from the two other MR Techniques aligned with the results obtained through the random-effects IVW approach. Sensitivity analysis between SS and vitiligo showed that the p-values of Cochran's Q statistics were all greater than 0.05, suggesting that there was no heterogeneity. The MR Egger intercept test indicates there was significant horizontal pleiotropy (p < 0.05). The MR-PRESSO methodology did not find the presence of outliers. The results of the sensitivity analysis between AS and vitiligo were as follows: Cochran's Q statistic showed no heterogeneity (p > 0.05). The MR Egger intercept test revealed no horizontal pleiotropy (p > 0.05). No outliers were identified in the MR-presso test. The aforementioned findings indicate that there is insufficient proof to establish a genetic causal connection between vitiligo and other rheumatic diseases (SS and AS). leave-one-out sensitivity analysis for vitiligo on Sjogren Syndrome and AS was shown in Figure S1. Sensitivity analysis of the MR analysis results in Table S3.
4 DISCUSSION
This study investigated the causative association between vitiligo and common rheumatic diseases, such as RA, SLE, SS, and AS. Using a two-sample MR analysis, we demonstrated that individuals with vitiligo were causally related to a higher risk of RA and SLE. However, no clear genetic causal relationship was found between vitiligo and other rheumatic diseases.
Many previous observational studies have found a high prevalence of RA in vitiligo patients, which is consistent with our findings. For example, in 2023, Lee conducted an observational study that revealed that patients with vitiligo show a notably increased risk of developing RA (OR = 1.82, 95% CI = 1.55–2.15).17 This positive connection is also demonstrated by a large amount of research investigating the association of vitiligo with autoimmune arthropathy using an inpatient database, which demonstrates that vitiligo is highly associated with an elevated risk of any form of inflammatory arthritis (OR = 2.23, 95% CI = 1.96–2.54), psoriatic arthritis (OR = 4.74, 95% CI = 2.97–7.56), RA (OR = 2.09, 95% CI = 1.82–2.41), and other inflammatory arthritis (OR = 4.14, 95% CI = 2.16–7.96).18
Our study found a significant association between vitiligo and SLE, which is also confirmed by previous studies. A large-scale epidemiological study of 86,210 vitiligo patients and 172 420 controls shows that vitiligo patients were at an increased risk of SLE (OR, 2.099; 95% CI, 1.478–2.982).19 In a systematic study, patients with vitiligo had increased odds of developing SLE (OR = 1.96, 95% CI = 1.52–2.52).17
In clinical practice, although there are many studies investigating the relationship between RA, SLE, and vitiligo, the exact reason for the association is not clear, and we analyze the possible reasons from the following aspects:
First, it is widely acknowledged that RA and SLE represent a chronic autoimmune disorder. However, the classification of vitiligo as an autoimmune disease remains relatively unknown among the general population.20 Autoimmune diseases are caused by the weakening or destruction of the body's autoimmune tolerance and the immune response of the body's immune system to its own tissues or components. The precise pathogenesis of vitiligo is still unclear and complex, but the autoimmune theory is the most widely acknowledged viewpoint.21 Therefore, the pathogenesis of RA, SLE, and vitiligo may involve common immunological factors. Individuals who have vitiligo display abnormal immune system activity, which may make them more susceptible to RA and SLE.
Second, the current data highlight that immune cells and their mediators play a pivotal role in the immunopathogenesis of vitiligo. Immune cells, such as helper T cell 1 (Th-1), cytotoxic T cell (CTL), and regulatory T cell (Treg), are the key immune cells.22 Among them, Tregs are vital for increasing peripheral immunological tolerance and defending the body against autoimmune injury.23 The accumulated data indicate that the number of Tregs is decreased in vitiligo patients, there is reduced inhibition of Tregs, and there are decreased levels of Treg-related inhibitory cytokines such as transforming growth factor-β.24, 25 Similarly, numerous autoimmune disorders, including RA, SLE, T1D, and multiple sclerosis, have been linked to abnormalities in the quantity and functionality of Tregs, which may be one of the reasons why vitiligo patients are susceptible to various related autoimmune diseases. About mediators, many key mediators, such as (IFN-γ) and C-XC chemokine ligand 10 (CXCL10), are also involved in the pathogenesis of vitiligo. A marker of Th1-mediated immune responses, CXCL10, was shown to be elevated in patients' serum with vitiligo.26 Recent research has demonstrated that the expression level of CXCL10 is increased in a range of organ-specific autoimmune diseases, including Graves' disease, autoimmune thyroiditis, T1D, and systemic rheumatic diseases such as RA, SLE, and systemic sclerosis, highlighting the significance of a common immune-pathogenesis of these diseases, which is characterized by a Th1 immune response.27, 28
Furthermore, the cytokine network is crucial to the pathophysiology of vitiligo, among which IFN-γ is one of the main cytokine networks.22 IFN-γ acts through the JAK/STAT pathway mediates cytokine signal transduction, induces innate and adaptive immune responses, and leads to JAK1-2 and STAT1 activation.29 Promising outcomes with JAK inhibitors in the treatment of vitiligo demonstrate the critical role of the JAK/STAT pathway in vitiligo.30-35 Likewise, the JAK-STAT pathway plays an important role in the pathogenesis of RA. The signal transduction of many key cytokines involved in the pathogenesis of RA is through the JAK-STAT pathway, which is a potential target for JAK inhibition.36
Finally, through genetic research, more than 50 vitiligo-related genes and loci have been identified. Interestingly, in genetic research on other autoimmune and autoinflammatory diseases, approximately 50% of the vitiligo susceptibility genes have been identified. This suggests that vitiligo and these diseases share a common autoimmune susceptibility.37
5 CONCLUSION
Our MR study provides evidence supporting the idea that vitiligo leads to a higher risk of RA and SLE. This finding emphasizes the need for patients with vitiligo to maintain a high level of vigilance and be aware of the possible risk of developing RA and SLE. Regular monitoring is essential to manage and address this potential complication. In addition, further validation of our findings in larger samples is warranted, and further in-depth studies are required to investigate the potential mechanisms of the cause-and-effect relationships described above.
ACKNOWLEDGMENTS
We appreciate the involvement of all participants and researchers in this study.
Open Research
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the Supplementary Material of this article.




