LncWNT3-IT affects the proliferation of Sertoli cells by regulating the expression of the WNT3 gene in goat testis
Lina He and Qijie He should be considered joint first author.
Contents
The proliferation and differentiation ability of testicular Sertoli cells directly affects spermatogenesis and male reproductive development. WNT proteins are involved in the regulation of cell proliferation, differentiation and spermatogenesis. Therefore, to study whether lncRNAs, which regulate the expression of WNT proteins during cell proliferation and differentiation, are worthwhile. In this study, testicular tissue from the Dazu black goat (Capra, goat, Chongqing, China) at neonatal time (less than 7 days old), early puberty time (45 days old) and sexual maturity time (90 days old) at three ages was subjected to high-throughput sequencing to predict testicular growth and development associated with WNT lncRNA. The final screening of lncWNT3-IT may be targeted to regulate the expression of WNT3. At the same time, the expression of WNT3 was verified by lncWNT3-IT by paraffin sectioning, fluorescence in situ hybridization, interference, overexpression, cytotoxicity assay, Western blotting and qPCR. The following results were obtained: lncWNT3-IT was expressed in the testicular Sertoli cells and played a role in the Sertoli cell cytoplasm. Fluorescence in situ hybridization localization analysis showed that lncWNT3-IT positively regulated the expression of WNT3, and through cell viability and cell proliferation experiments, it was found that the expression of lncWNT3-IT assisted in Sertoli cell proliferation. In summary, lncWNT3-IT can influence the proliferation of Sertoli cells by positively regulating the expression of WNT3.
1 INTRODUCTION
Sertoli cell proliferation and differentiation directly affect spermatogenesis in male reproductive development. Studying the molecular mechanism of the Sertoli cell growth and development, exploring the key factors regulating their proliferation and differentiation, and laying a foundation for studying their function are of great significance for the improvement of the production level and sperm quality of rams (Savvulidi, Ptacek, Vargova, & Stadnik, 2019; Wang, Xia, Rehm, & Fan, 2015).
The term WNT arises from the contraction of the Wingless gene name (Wingless-related MMTV integration site), which was first reported by Nusse and Varmus in 1982 (Nusse & Varmus, 1982). WNT3 is a member of this protein family and affects the critical stages of development and differentiation by activating the typical signalling pathway WNT/β-catenin (Weng et al., 2017). In recent years, new studies have found that the expression of WNT3 in adolescent monkeys Sertoli cells (active spermatogenesis) is significantly higher than that in young monkeys (spermatogenesis) (Basu et al., 2018), and studies in mice have indicated the same. The low expression of the Sertoli cells in WNT3 reduces the expression of connexin 43, a linker molecule essential for germ cell development, indicating that WNT3 expression impairs the Sertoli cell-mediated spermatogenesis and affects male fertility (Basu et al., 2018; Kerr, Young, Horvay, Abud, & Loveland, 2014; Lopez et al., 2019). Abnormal WNT signalling is one cause of animal and human pathology, including testicular (Boyer, Paquet, Lague, Hermo, & Boerboom, 2009; Chang, Guillou, Taketo, & Behringer, 2009) and prostate cancer (X. G. Li et al., 2019; Niu, Hou, & Li, 2018; Sun & Liu, 2017).
Other studies have shown that long non-coding RNAs (lncRNAs) play a key regulatory role in many important biological processes, by participating in oocyte maturation (J. Li et al., 2015), sperm formation (Anguera et al., 2011), formation of the placenta (Zheng et al., 2013), pregnancy (Nakagawa et al., 2014), gonadal hormone response (W.B. Li et al., 2013), embryonic development (Durruthy-Durruthy et al., 2016) and sex determination (Mulvey, Olcese, Cabrera, & Horabin, 2014). More studies have shown that lncRNAs play an important role in the regulation of mammalian spermatogenesis, including a series of processes during sperm cell proliferation, spermatocyte meiosis and morphological changes of round sperm. Furthermore, lncRNAs affect sperm maturation in a dynamic expression pattern (Bao, Wu, Schuster, Hennig, & Yan, 2013; Liang et al., 2014; Luk, Chan, Rennert, & Lee, 2014). These RNAs may partially play a regulatory role in spermatogenesis. While studying mouse chromosome 8, Ganesan and Rao found a new lncRNA, mrhl (meiotic recombination hot spot locus, mrhl) (Ganesan & Rao, 2008). The study found that mrhl RNA might be regulated by two molecular mechanisms of spermatogenesis (Akhade, Arun, Donakonda, & Rao, 2015; Arun, Akhade, Donakonda, & Rao, 2012). Mrhl is the first lncRNA to be identified as involved in the regulation of WNT signalling (Kerr et al., 2014). This implies that the lncRNA influences on the WNT signalling regulatory mechanism may have a very important role in spermatogenesis.
In our previous study, we screened testicular development in relation with the lncRNAs by high-throughput sequencing (He, Zhao, Dai, Qiao, & Yang, 2019). The multiomics analysis revealed that one of the lncRNAs coincided with the antisense strand of the WNT3 gene and we named it lncWNT3-IT (Intron lncRNA). The target gene function of lncWNT3-IT is enriched in the relevant pathway of proliferation of the Sertoli cells, which is similar to the function of WNT3. Therefore, we hypothesized that lncWNT3-IT regulated proliferation of the testicular Sertoli cells via WNT3. In this experiment, we used the Dazu black goat as experimental subject to explore this hypothesis at various levels, by considering different age groups, tissues, cells, cellular localization and expression.
2 MATERIALS AND METHODS
2.1 Animals
The experimental conditions were approved by the Committee on the Ethics of Animal Experiments of Southwest University, China (No. [2007] 3) and the Animal Protection Law in China, and all efforts were made to minimize animal suffering.
Experimental animals were obtained from the Dazu Black Goat Farm at Southwest University, Chongqing, China. As a good local variety in Chongqing, China, the Dazu Black Goat had the characteristics of good reproductive performance and early sexual maturity. The goats selected in the experiment were strong, well-nourished and had no reproductive-related diseases. When any goat was born, the rams with better health and normal testicular development were selected and raised under the same feeding conditions (house feeding, without restriction of feed and water). From each of the following age group, 3 rams were selected: less than 7 days old (neonatal time, NT), 45 days old (early puberty time, ET) and 90 days old (sexual maturity time, ST), for a total of 9 rams. After the goats were reared to the respective sampling age, their testes were collected after local anaesthesia. The collected testes were stored in two parts. One part was disinfected with alcohol and placed in the laboratory at 32°C in a physiological saline solution containing 0.25% streptomycin for separation and culture of the Sertoli cells. The other part was used in the fluorescence in situ hybridization (FISH) experiments. After alcohol disinfection, the tin foil package was quickly placed in liquid nitrogen and brought back to the laboratory for storage at −80°C.
2.2 Culture and identification of the Sertoli cells
After the testicles were sterilized in 75% alcohol for 30 s, they were washed three times with PBS buffer. The surface PBS was removed with pure DMEM/F12 medium (Gibco, USA), and the samples were then placed in a sterile Petri dish to remove the testicular tunica albuginea. Visible blood vessels and connective tissue were cut. A small amount of testicular parenchyma was cut and washed twice with PBS; then, the PBS was washed off the parenchyma surface with pure DMEM/F12 medium. The tissue was cut into 1 mm-2 mm pieces with ophthalmic scissors and transferred to a 50-ml centrifuge tube. Five volumes of 4 mg/ml collagenase IV (Thermo Fisher Scientific, USA) and 0.2% hyaluronidase (Thermo Fisher Scientific, USA) were mixed, digested and placed in a 37°C constant temperature shaker for 30 min. The samples were then centrifuged at 250 g for 5 min, the supernatant was decanted, and DMEM/F12 medium containing 14% foetal bovine serum (ExCell Bio, Australia) was added; the samples were centrifuged again at 250 g for 5 min and then washed twice. The supernatant was decanted, mixed with 5 volumes of 0.25% trypsin (Thermo Fisher Scientific, USA) and 10 μg/ml DNAse I for the digestion, placed in a 37°C constant temperature shaker for 15 min and shaken several times. An equal volume of foetal calf serum was added to terminate the digestion; then, the culture was sieved through 80-mm and 300-mm stainless steel sieves to obtain a single-cell suspension. The cells were centrifuged at 250 g/min for 5 min; then, 5 ml doses were seeded to culture flasks containing 14% foetal bovine serum (ExCell Bio, Australia) by uniform pipetting. After 2 hr, the solution was discarded, and the cells were washed with PBS buffer, suspended in a new 14% foetal bovine serum solution and cultured at 37°C with 5% CO2. Primary cells were cultured for 3 days and transferred when the culture grew to more than 80% of the bottom of the flask. The cells were inspected for cell growth every 24 hr after passage, and subsequent experiments were performed after stable growth was observed (48 hr-72 hr).
Sertoli cells isolated from the testis and cultured in vitro needed to be identified for subsequent experiments. In order to identify the quality and purity of the cultured Sertoli cells, we detected the specific expression of WT1 in the Sertoli cells by immunofluorescence staining. Owing to this, during the FISH overnight incubation at 4°C with the primary antibody, 50 μL of WT1 primary antibody (1:250; Abcam trading co., LTD, Shanghai) was added to each slide. During the 2 hr incubation at room temperature with the secondary antibody, fluorescein isothiocyanate (FITC, excitation light wavelength: 519 nm) was added to mark the goat anti-rabbit secondary antibody (Abcam trading Co., LTD).
2.3 LTCONS_00052549 positioning
To explore the mechanism of action of lncRNAs, it was necessary to localize them in the cell and study their action site, which laid a foundation for the subsequent study of their regulation mechanism. The probe sequence used for this was 5′-TGAGAAAACCAGGTCTCTGAGCTTATTATCCCTTCAAGTCTGTGTATA-3′
2.3.1 Tissue section FISH
For ease of preservation, testis tissue acquisition was followed by paraffin sectioning. The paraffin sections were dewaxed in xylene until the wax was completely removed; then, the tissue sections were removed, placed twice in anhydrous ethanol, each time for 5 min, and dried at room temperature. Proteinase K digestion at 55°C for 10 min was followed by two washes in 2X SSC, each time for 5 min. Pre-cooled (−20°C) 70%, 85% and 100% alcohol dehydration for 5 min was followed by air drying. Sections were immersed twice in a pre-heated (78°C) denaturing solution for 8 min. An appropriate amount of pre-hybridization solution was added to the sample, covered with membrane and left to stand for 30 min. In the pre-hybridization, the CY3 fluorescent dye-labelled probe (Guangzhou, China) and the hybridization solution were mixed into a probe hybridization mixture, denatured at 78°C for 8 min and quickly placed on ice for use. The hybridization cocktail was added to the sections for overnight incubation at 42°C. Then, 50% formamide in 2X SSC was used to wash the membranes twice at 53°C, each time for 5 min, followed by a 42°C wash with 0.1% NP-40/2X SSC twice, each time for 5 min. Next, the membranes were washed with 0.5X SSC, 0.2X SSC and 0.1X SSC at 42°C, twice for 5 min for each solution, then with 1X PBS at room temperature(25℃) for 15 min.
2.3.2 Cell slide FISH
The cells were washed with PBS before the experiment. The cell slides were fixed with 4% paraformaldehyde for 20 min-30 min. We then washed the samples three times in distilled water for 5 min. Permeated 0.5% Triton X-100 was added for perforation for 20 min, followed by two 5 min washes with 2X SSC. Samples were turned into pre-cooled (−20°C) 70%, 85% and 100% dehydrated alcohol, 5 min each, and air-dried. Next, we used a hybridization solution and probe mix for 73°C denaturation for 5 min-8 min and then on ice for 5 min. We added the samples to a mixture of hybrid cells crawling on-chip, followed by overnight hybridization at 37°C. Samples were washed with pre-warmed (43°C) 50% formamide with 2X SSC for 5 min (three times) and 2X SSC for 5 min (two times). They were then stained with DAPI counterstaining for 5 min and washed twice with 2X SSC. Anti-decay fluorescent mounting medium (Shanghai, China) was used for mounting and observation under a fluorescence microscope.
2.4 Total RNA, reverse transcription and real-time PCR
Total RNA was extracted using the TRIzol reagent (Tiangen, China) according to the manufacturer's instructions and transferred to Huada Gene Company for quality integrity testing. First-strand cDNA was synthesized according to the instructions of the Genomic cDNA Kit using the FastKing one-step method. A quantity of 50 ng-2 μg total RNA was used to establish a 20-μL reaction system for qPCR performed using the SsoAdvanced™ SYBR Green Supermix (Bio-Rad) and the CFX96™ Real-Time System (Bio-Rad) according to the manufacturer's instructions. The reference nucleotide sequence was queried in the NCBI database according to the species and name of the gene of interest. All genes were designed with the primer design software Primer Premier 5.0 (Premier, Canada). The primer sequences were commissioned to Biotech Engineering (Shanghai) Co., Ltd. The internal reference gene was either β-actin or GADPH, for different analyses. The primer sequences have been shown in Table 1. Data were analysed using the 2−ΔΔCT method.
| Gene | Primer sequences (5′-3′) | |
|---|---|---|
| WNT3 | F | TAGTTCTCCAGGGTCTGATAGG |
| R | CTGTTGGAGTAGAAGTGGGTTT | |
| lncRNA | F | TGAGATCAAGAAGGTGGTGAAG |
| R | GCATCGAAGGTAGAAGAGTGAG | |
| β-actin | F | CCTGGACTTCGAGCAGGAGA |
| R | AGGCTGGAAGAGAGCCTCAG |
Note
- F, forward primer; R, reverse primer.
2.5 Construction and identification of overexpression vector and interference fragment
To verify the effect of lncWNT3-IT on WNT3, we designed overexpression vector processing and interference fragment processing. The sequence of LTCONS_00052549 has been shown in Table 2, and the vector map has been shown in Figure 1. Positive colonies were identified by PCR and sent for sequencing to the Huada Gene Company. Endotoxin-free plasmid extraction was performed. The three pairs of interference fragments were designed and synthesized by Biotech Engineering (Shanghai) Co., Ltd according to the sequence of LTCONS_00052549, and the negative control (NC) was a sequence with no homology in mammals. The interference sequence fragments have been shown in Table 3.
| LTCONS_00052549 sequence |
|---|
| 5′-CCCGCCTTCGCCGCCTCGCGCCTTCCGGCCGCCGACCCCGGCGCCCCCTGCCGCCGCTCTGGGCGCGCCGGCCCCGGGGAGTCGGTCCCCGGCGGCCGCGGAAGATGCGCGAGCCCCGGCCCAGACCGCCTCCCTGCCACCCTGGGTCTCAAATCCGGGGGCAGAGGTCTACCCTGGCAGCCCGCCCGGCGCGGGTAGGGGGCTCGCCAGAGGCGGGGCGCGAGCGTGGAGCCCATAGCGGGGTTCGAACCCGTTCACGCCAGGCTTCGGGCACTGGGGAGCGCGTGCGGGCGCGGGGTGGGGGGGCCTCCTCCCGCAGACCGCTGAGTTCCCCTTTCTCTGACTTGGGAACCTGAGCTCGCTCACGTTCCTTTCTCTGACATCCTACCCACGCCTCCGCATCACAAAACTGTCCTGCAATTTCGGGATGTGGGCAAGTCCTAGAACCTACAACGGGGATTTAAAACGTTATACACAGACTTGAAGGGATAATAAGCTCAGAGACCTGGTTTTCTCAGGTTTTCACATCCCGCGCGGAGAAGAGCAATCAGAAGGCCAAGCCTCTTCCTCGGATCTGCCTCATTTAGTCCTTGTAACAACTCCAGGACCTAGGAGGGTTGTTACAGCAGCTCGTTTCCCATGACTTAGCCAGACAGGTGGAGTAACCAACCCAGACCACAGAGCTACCCAAGGGATGGCTTACTGGAACCTGTTCACCTGATTCCAGAGCACGTGGGGCACGTACCACCCTCCTCTGGGCAGTGCCTGCCTGGGCCCTGCAGGGTAGGAGTGGCTGCAGGCCTGGCCCGTCTGCACCCTGGGCCTCTAGACTGAACCCAGGTGATAGGGTGGGTCGGCCCAGAGCCCCACGATGGGCACCAGGACGCACACAGCATCTGCTGGGTTGCCCTCCCTGCCTCTGCCATTTACAGTTATAAACTGGTTATTGTGACAATAAT-3′ |
| Type | Number | F (5′-3′, overhangs included) | R (5′-3′, overhangs included) |
|---|---|---|---|
| siRNA1 | S144 | AAAUUGCAGGACAGUUUUGUGUU | CAAAACUGUCCUGCAAUUUCGUU |
| siRNA2 | S145 | UUUAAAUCCCCGUUGUAGGUUUU | CCUACAACGGGGAUUUAAAACUU |
| siRNA3 | S146 | UUUUAAAUCCCCGUUGUAGGUUU | CUACAACGGGGAUUUAAAACUU |
| siRNA NC | S20 | UUCUCCGAACGUGUCACGUdTdT | ACGUGACACGUUCGGAGAAdTdT |
Abbreviation:
- F, forward primer; R, reverse primer; S144, S145 and S146, three interference segments of LTCONS_00052549 sequence; siRNA NC, a sequence that has no homology to mammals.
The siRNA was transfected into the Sertoli cells. The day before transfection, the cells were digested with trypsin and then planked, and the cell confluence on the second day was controlled to be between 80% and 90%. Transfection reagent, siRNA and serum-free DMEM were balanced to 15°C–25°C. The siRNA was diluted with serum-free DMEM medium, and the EndoFectin-Lenti reagent (HyClone) was diluted with the same medium. The siRNA solution was mixed gently, enriched with drops of the diluted transfection reagent mixed again and allowed to stand at room temperature for 10 min-25 min to form the DNA-transfection reagent complex. Drop by drop, the DNA-transfection reagent complex was added to the Petri dish while mixing, then transfected and cultured in an incubator at 32°C and 5% CO2. After 24 hr–48 hr, the cells were collected for experimental detection.
2.6 Extraction of total proteins in the Sertoli cells and Western blotting
First, the cell culture medium was discarded, and the suction flask was washed twice and cleaned with PBS buffer. Trypsin (2 ml) was added to the bottles; then, the bottom of the culture flask was repeatedly mixed and vigorously tapped to make all adherent cells float. The cells and their medium containing foetal bovine serum were transferred to 2-mL centrifuge tubes. The cell suspension was centrifuged at 700 g for 5 min. The supernatant was discarded; 1 ml PBS buffer was added, moved into a 1.5-mL centrifuge tube, centrifuged at 4°C, 10,000 g for 5 min and washed twice; the procedure was repeated two times overall. Then, we added 1 ml of the cell lysate (containing phenylmethylsulfonyl fluoride, 1:200), repeated the mixing, lysed on ice for 30 min, centrifuged at 4°C at 10,000 g for 10 min and transferred the supernatant to an EP tube. The loading buffer was added at a 4:1 ratio to the protein, mixed and boiled for 2 min to denature the protein. A separating gel and a stacking gel were prepared for use. We then performed electrophoresis, transfer and blocking. An anti-specification reference, an antibody dilution (1:1,000), and the PVDF membrane were placed in each antibody solution and incubated overnight at 4°C. We then added TBST washing solution, shook and washed three times for 15 min. The reference specification secondary antibody and diluted horseradish peroxidase (HRP) (Abcam (Shanghai) trading Co., LTD) were mixed, and the PVDF membrane was added to it (1:5,000) and agitated for 1 hr at room temperature. With the same procedure, an antiwash was executed with the BeyoECLPlus chemiluminescence reagent to develop colour in the dark, and the sheet was compressed with an X-ray film developing solution and fixing solution.
2.7 Cell proliferation activity and cycle detection
We used the CCK-8 kit (Beyotime Bio) to detect cell proliferation according to the manufacturer's instructions. It is a quick and highly sensitive detection kit based on WST-8, which is widely used for cell proliferation and cytotoxicity. WST-8 is a compound similar to MTT (employed in the classical MTT cell proliferation assay) and can be reduced by some dehydrogenases in the mitochondria to produce orange formazan in the presence of an electron-coupled reagent. For cell cycle detection, intracellular DNA can be combined with a fluorescent dye (such as propidium iodide). The DNA content of the cells varied at different times, with different fluorescent emission. The fluorescence intensity detected by flow cytometry was also different. Therefore, cells can be observed by measuring changes in cell light scattering by flow cytometry.
2.8 Statistical analysis
Three replicates were set for each set of experiments; the results were expressed as ‘mean ± SEM (standard error of the mean)’ and analysed using one-way ANOVA. All results were considered statistically significant at p < .05, and statistical analyses were performed using the Statistical Product and Service Solutions (SPSS) software 18.0 (SPSS, Inc).
3 RESULTS
3.1 Sertoli cell collection and identification
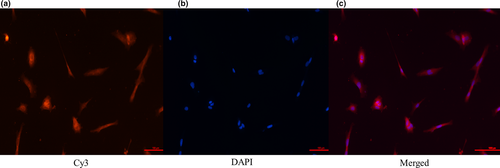
The results showed that the Sertoli cells specifically expressing WT1 were stained red and the nuclei were stained blue with DAPI. Through comparison, it was found that the cultured cells in this experiment were indeed highly pure testicular Sertoli cells of the Dazu black goat that could be used in subsequent experiments. (Figure 2).
3.2 Expression profile of lncWNT3-IT and WNT3 in the three age groups
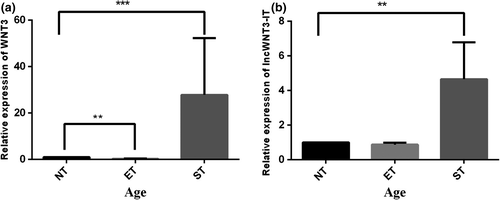
To detect whether lncWNT3-IT was present, we designed and synthesized lncWNT3-IT specific primers, and both undertook quantitative fluorescence experiments and calculated expression profiles for WNT3 and lncWNT3-IT in three age groups. It could be observed that lncWNT3-IT expression and WNT3 expression showed a consistent trend in the three age groups: their expression in the ET and ST groups was higher than those in the NT subjects (p < .01, Figure 3).
3.3 Positioning, interference, and overexpression design and screening of lncWNT3-IT
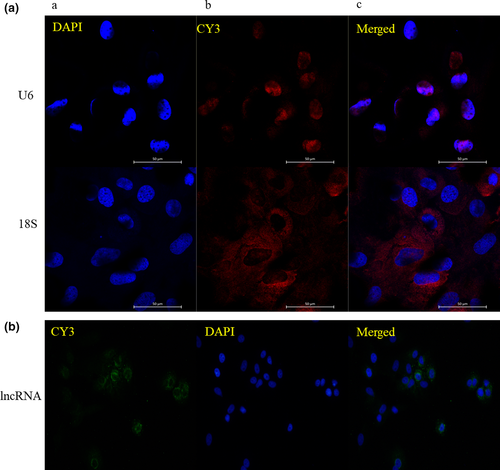
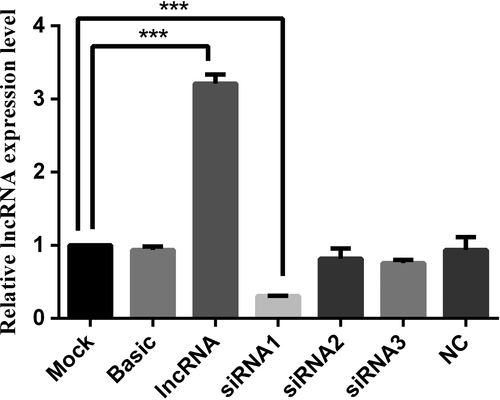
To more accurately investigate the mechanism of action of lncWNT3-IT, we used FISH experiments (Figure 4) to determine its nuclear distribution in the Sertoli cells. FISH results showed that lncWNT3-IT was abundant in the cytoplasm of supporting cells, but only a few positive signals were detected in the nucleus. A total of three sets of interference fragments, named siRNA1, siRNA2 and siRNA3, were designed and synthesized with an overexpression vector. Fluorescence quantitative detection of the three pairs of interfering fragments and their overexpression efficiency is shown in Figure 5). In the end, siRNA1 was selected for subsequent experiments.
3.4 LncWNT3-IT positively regulates the expression of WNT3
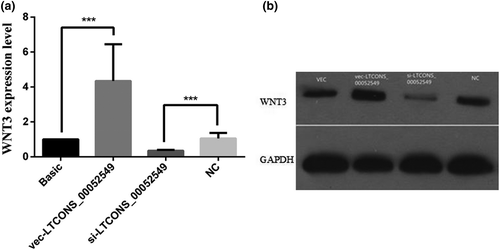
To examine the effect of lncWNT3-IT on WNT3 gene expression levels, we first designed a set of experimental groups and transfected the cells with the interfering construct lncWNT3-IT, based on the selected siRNA1, in differential expression vectors. The four constructs were as follows: the overexpression vector negative control, with no silencing sequence included (vec group), the vector for the overexpression of the lncWNT3-IT (vec-LTCONS_00052549 group), a normal vector containing the silencing sequence serving as a negative control (NC group) and the interfering vector containing the silencing sequence (si-LTCONS_00052549 group). After the transfection, WNT3 expression decreased (p < .001) in the silenced group, while it increased in the overexpression group (p < .001) compared to that in the NC group. This indicates that the increase or decrease in lncRNA expression can significantly affect the expression level of WNT3 (Figure 6a). LncWNT3-IT positively regulates WNT3 gene expression. Western blotting results showed a similar pattern (Figure 6b).
3.5 LncWNT3-IT affects cell viability and cell cycle
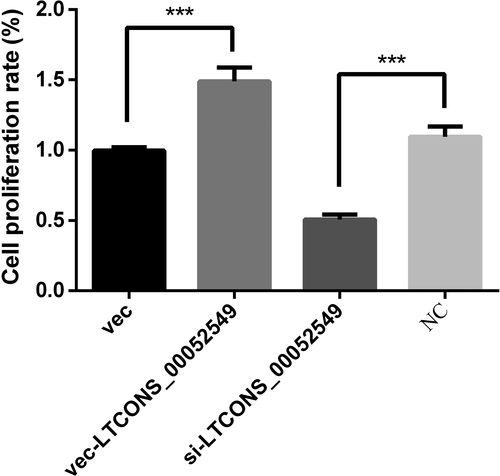
To determine the effect of lncWNT3-IT on the Sertoli cell viability, we used the CCK-8 kit on cells transfected with the same four constructs based on silencing sequences and differential expression vectors. The results showed in Figure 7 prove that the transfection of Sertoli cells increased cell viability in the vec-LTCONS_00052549 group compared to the NC group (p < .001), while cell viability decreased in the silenced group (p < .001). This showed that the expression level of lncWNT3-IT directly influenced the level of cell activity by promoting cell proliferation when present and inhibiting cell proliferation when absent.
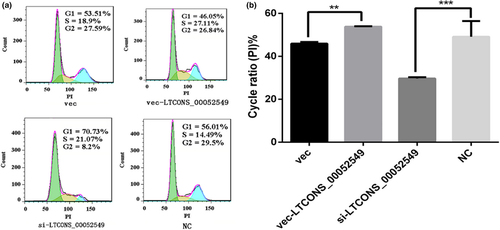
To quantify the positive effect of lncWNT3-IT on the growth phase, the transfected cells were cultured with a cell cycle kit and counted by flow cytometry. The results showed that, compared with the NC group, the si-LTCONS_00052549 group had a decrease in the cell proliferation index (p < .001) in the G0/G1 phase, while the vec-LTCONS_00052549 group showed an increase in the cell proliferation index in the S + G2/M phase (p < .01) compared to the vec group (Figure 8). The results indicate that a higher expression level of lncWNT3-IT promoted cell proliferation, while reduced lncWNT3-IT expression inhibited it.
4 DISCUSSION
4.1 LncWNT3-IT affects the expression of WNT3
Through expression analysis in the Sertoli cells of three age groups, it was found that lncWNT3-IT and WNT3 had similar expression trends during the differentiation of Sertoli cells; that is, with the process of Sertoli cell differentiation and maturation, both the expression levels significantly improved, suggesting that they may play an important role in the regulation of cell proliferation and differentiation (Basu et al., 2018; Coombs et al., 2010). It is common knowledge that the vast majority of the Sertoli cells cease proliferation at puberty, when the blood–testis barrier is established and spermatogenesis starts. Cessation of proliferation is a prerequisite for the Sertoli cell maturation and for the establishment of the blood–testis barrier (Meroni et al., 2019). There are quite recent data suggesting that some Sertoli cells may proliferate in the adult rodent (Fig ueiredo, Franca, Hess, & Costa, 2016). In this study, high levels of lncWNT3-IT and WNT3 were expressed in the Sertoli cells during sexual maturation. Studies have shown that WNT3 can promote cell proliferation through the β-catenin pathway (Golestaneh et al., 2009). In a study on non-human primates, WNT3 was the only WNT ligand and the testicular Sertoli cells in adolescence continued to increase. Additionally, the expression of WNT3, a morphogen of the WNT/β-catenin pathway, was higher in the pubertal Sertoli cells than those in the infant Sertoli cells (Basu et al., 2018). This is similar to the results of this study. LncWNT3-IT showed a similar trend to that of WNT3 during testicular tissue growth and development; that is, with testis maturity, the proliferation and differentiation of the Sertoli cells, and the occurrence of spermatogenesis, the expression levels of both were significantly higher to support post-testicular development and spermatogenesis, as cell proliferation plays a regulatory role in them. The lncWNT3-IT is highly likely to promote the expression of the WNT3 gene by accumulating in the vicinity of the WNT3 gene promoter, thereby regulating the protein translation of WNT3 in a cis-acting manner; this may exert a post-transcriptional regulation function, increase the expression of WNT3 protein, and promote proliferation and differentiation of the Sertoli cells. This has a mechanism similar to what was observed in previous studies on the lncRNA intron (Basu et al., 2018; Yin et al., 2012). In relation with the study by Zhang et al., we suggest that lncWNT3-IT has a mechanism of action specular to antisense lncRNA: lncWNT3-IT may regulate the expression of WNT3 by accumulating at the promoter end of the WNT3 gene (Zhang et al., 2013). Localization analysis also showed lncWNT3-IT accumulated in the cytoplasm of the Sertoli cells. However, some antisense lncRNAs with the same expression tendency as the coding gene (Wei et al., 2015) were found to inhibit the translation process of the protein by complementing the mRNA in the post-transcriptional phase. To verify whether the lncWNT3-IT intron and antisense lncRNA behaved similarly, inhibition experiments with WNT3 protein translation and protein levels in different treatment groups were tested; the results showed similar effects. It has been shown that the intron lncWNT3-IT can promote the transcription process of the WNT3 protein in the cytoplasm, while positively regulating the transcription process of the gene in the nucleus (Figure 9).
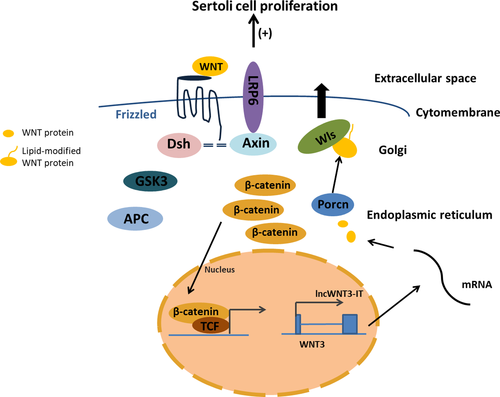
4.2 Effect of lncWNT3-IT on the Sertoli cells
We verified that changes in the expression of WNT3 caused by changes in the expression of lncWNT3-IT could also affect the proliferation of the Sertoli cells. LncWNT3-IT upregulated the proliferation of the goat Sertoli cells (p < .001) and influenced their growth cycle. This indicates that lncWNT3-IT can regulate the proliferation of the Sertoli cells through the expression of WNT3. This result is consistent with the results we initially hypothesized. The results of cell cycle experiments showed that the expression level of WNT3 increased after the expression level of lncWNT3-IT increased in the group transfected with the overexpression vector. In the S + G2/M phase, the Sertoli cells in the vec-LTCONS_00052549 group had a higher cell proliferation index compared with the vec group. In the Sertoli cells transfected with siRNA group, both lncRNA and WNT3 expression decreased, while the cell cycle was comparable to the one observed in the NC group: the cell cycle arrested at the G0/G1 phase, and the proliferation index decreased significantly. Similar to the phenomenon observed by Basu et al. (Basu et al., 2018) in WNT3 knockdown mice, we found that decreased expression of WNT3 resulted in suppression of proliferation and differentiation of the Sertoli cells. This results in an arrest of the cell cycle at the G0/G1 phase. Furthermore, lncWNT3-IT increased the persistence of the S + G2/M cell phase: the G0/G1 phase was much more frequent than the S + G2/M phase, and the molecular activity reflected this.
In summary, it is suggested that the process of lncWNT3-IT affecting the proliferation of the Sertoli cells is likely to affect the WNT3 promoter activity, thus regulating the gene expression and protein translation process, affecting WNT3 protein levels. Additionally, WNT3 can regulate the synthesis of the cell division-related proteins such as chromatin proteins and DNA helicase, that support the G0/G1 phase (pre-DNA synthesis), thereby affecting the proliferation and differentiation of the Sertoli cells. The mechanism of action of lncWNT3-IT in regulating WNT3 expression and the molecular mechanism of WNT3 affecting the proliferation of the Sertoli cells also needs to be explored and verified.
5 CONCLUSIONS
In summary, the intron lncRNA WNT3-IT in the testicular Sertoli cells of the Dazu black goat can regulate the proliferation of Sertoli cells in the G0/G1 phase by positively regulating the expression of WNT3 in the Sertoli cells. Proliferation and differentiation of Sertoli cells play an important role in late testis development and spermatogenesis. This study provides fundamental data for the study of Sertoli cells and lays a foundation for the exploration of the molecular regulation mechanism of WNT3 expression conducted by lncWNT3-IT.
ACKNOWLEDGEMENTS
We gratefully acknowledge Dr. Lingbin Liu for the critical reading of this manuscript. This work was supported by the National Key Research and Development Program (2018YFD0502003) and the Chongqing Municipal Technology Innovation and Application demonstration special social livelihood key research and development program (cstc2018jscx-mszdx0044).
CONFLICT OF INTEREST
None of the authors have any conflict of interest to declare.
AUTHOR CONTRIBUTION
Lina He wrote the manuscript; Qijie He designed the experiment; Lei Qiao and Siyi Huang conducted experiments; TianyuanYang and Zinuo Dai conducted data analysis; Lingbin Liu critically reviewed the manuscript; and Zhongquan Zhao was the instructor of this experiment.
Open Research
DATA AVAILABILITY
The data that support the findings of this study will be available from the corresponding author upon a reasonable request.




