MoCloro: an extension of the Chlamydomonas reinhardtii modular cloning toolkit for microalgal chloroplast engineering
Abstract
Photosynthetic microalgae are promising green cell factories for the sustainable production of high-value chemicals and biopharmaceuticals. The chloroplast organelle is being developed as a chassis for synthetic biology as it contains its own genome (the plastome) and some interesting advantages, such as high recombinant protein titers and a diverse and dynamic metabolism. However, chloroplast engineering is currently hampered by the lack of standardized cloning tools and Design-Build-Test-Learn workflows to ease genomic and metabolic engineering. The MoClo (Modular Cloning) toolkit based on Golden Gate assembly was recently developed in the model eukaryotic green microalgae Chlamydomonas reinhardtii to facilitate nuclear transformation and engineering. Here, we present MoCloro as an extension of the MoClo that allows chloroplast genome engineering. Briefly, a Golden Gate-compatible chloroplast transformation vector (pWF.K.4) was constructed, which contains homologous arms for integration at the petA site in the plastome. A collection of standardized parts (promoters, terminators, reporter and selection marker genes) was created following the MoClo syntax to enable easy combinatorial assembly of multi-cassettes in the destination pWF.K.4 vector. The functionality of the biobricks was assayed by constructing and assessing the expression of several multigenic constructs. Finally, a generic vector pK4 was constructed for easy Golden Gate cloning of 5′ and 3′ homologous arms, allowing targeting at alternative plastome integration sites. This work represents a significant advancement in technology aimed at facilitating more efficient and rapid chloroplast transformation and engineering of green microalgae.
1 INTRODUCTION
Green algae are emerging as promising biotechnological platforms for the sustainable production of bioproducts of interest, such as proteins, lipids, sugars, pigments, alcohols and other bulk chemicals. Microalgae are unicellular algae that possess some intrinsic advantages as green cell factories, as they can grow on atmospheric CO2 and sunlight without using agricultural land (Gangl et al., 2015; Dehghani et al., 2020). The eukaryotic green microalgae Chlamydomonas reinhardtii (from now on Chlamydomonas) is being developed as a model organism for synthetic biology (SynBio) and molecular biotechnology (Scaife et al., 2015; Schroda and Remacle, 2022). Chlamydomonas offers several advantages, such as a well-characterized genome, easy and inexpensive cultivation, and availability of sophisticated genetic manipulation tools. Indeed, Chlamydomonas has been used as a proof of principle to produce recombinant proteins, biodiesel, valuable metabolites and vaccines, among others (Rasala et al., 2011; Georgianna and Mayfield, 2012; Scranton et al., 2015; Shamriz and Ofoghi, 2016; Lauersen, 2019; Jarquín-Cordero et al., 2020).
The ability to introduce and express genes of interest in Chlamydomonas is crucial for advancing research and applications. Nuclear genome transformation has been a conventional approach, aided by technological advances addressed to overcome the strong gene silencing machinery that is present in Chlamydomonas (Neupert, Karcher and Bock, 2009; Baier et al., 2018; Crozet et al., 2018). Recently, chloroplast genome (the plastome) transformation has gained attention as it might induce higher levels of recombinant gene expression due to high plastome copy number, stable insertion at targeted plastome sites by homologous recombination and no transgene silencing machinery (Dyo and Purton, 2018). In contrast to higher plants, Chlamydomonas cells contain a single chloroplast. The number of plastome copies per chloroplast ranges from about 40 to about 100, depending on the physiological state of the cell (Jackson et al., 2021). Chlamydomonas plastome is around 205 kb in size with the typical structure of two inverted repeat regions that separate two single-copy regions, and it contains about 20% of repetitive DNA and encodes 108 genes (Maul et al., 2002; Jackson et al., 2021). In addition, chloroplasts have a very active and dynamic metabolism as they are the cellular compartments where photosynthesis, carbon assimilation, and biosynthesis of several essential metabolites take place. Recent reports in Chlamydomonas and other plant systems such as tobacco reinforce the interest of chloroplast as a compartment for metabolic engineering purposes, with the goal of producing valuable metabolites such as carotenoids, polyamines, terpenoid-derived drug artemisinin, or lipid precursors of rhamnolipid biosurfactants (Kaushal, Abdin and Kumar, 2020; Perozeni et al., 2020; Einhaus et al., 2022; Miró-Vinyals et al., 2023). Recombinant protein expression in the chloroplast also presents some challenges, such as no protein glycosylation, potential improper folding, no secretory system, or low transcription factor titer relative to the multiple plastome copies, possibly limiting transgene expression.
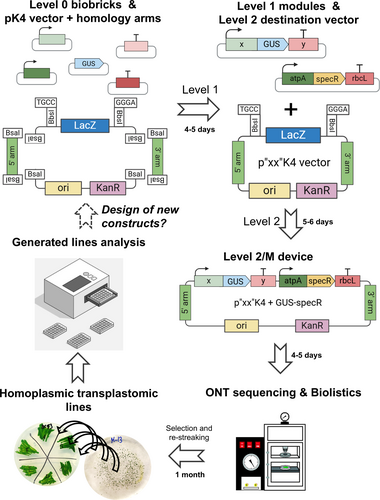
SynBio uses biology and engineering principles to design new biological systems for practical applications. The need for fast advancement in SynBio facilitated the incorporation of Design-Build-Test-Learn protocols requiring modular construction of parts called “biobricks”. A universal standard syntax was created for the design of biobricks by the OpenPlant Consortium, an international plant synthetic biology community (Patron et al., 2015). The syntax is based on Golden Gate Modular Cloning (MoClo), which uses Type IIS restriction enzymes that allow cutting DNA sequences at specific sites outside of the recognition site, therefore generating complementary overhangs that can be specifically designed for the modular assembly of the parts (Figure 1A). MoClo strategy allows the assembly of multigene constructs for any eukaryotic system (used for up to 11 transcription units) with high efficiency (Weber et al., 2011).
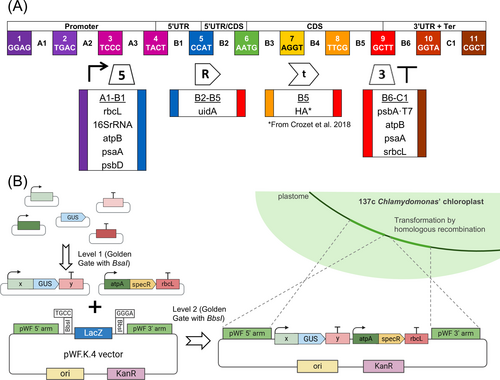
Despite the scientific and industrial interest in microalgae, it was just very recently that modern cloning techniques for easy genetic manipulation were established. A MoClo toolkit, following international standard syntax, was designed for Chlamydomonas nuclear transformation (Crozet et al., 2018). Using type IIS endonucleases, specific overhangs are generated for each piece (for example, sequences A1-B1 for promoters and 5′-UTRs or B2-B5 for coding sequences, Figure 1A), allowing directional assembly in a single Golden Gate restriction/ligation reaction. This syntax is defined by Level 0 plasmids containing standard gene parts (biobricks for promoters, coding sequences, untranslated regions, tags, etc.), and establishes specific overlapping sites upon restriction with BsaI for pre-determined linear cloning of up to 10 different positions (Figure 1A). In a single reaction tube, standardized parts can be assembled into modules (Transcriptional Unit, TU, Level 1) in standardized Level 1 backbone vectors (Weber et al., 2011). Then, in a second reaction using a distinct restriction endonuclease Type II, BbsI, strategically located in Level 1 destination vectors, modules are assembled into devices which can be longer than 10 TUs (multigenic construct, Level M or 2, Figure 1B).
Whereas nuclear MoClo provides a standardized methodology with successful implementation in Chlamydomonas SynBio (Freudenberg et al., 2022; Kiefer et al., 2022), standard biobricks and standardized vectors for chloroplast transformation are still missing. Given the interest in chloroplast genome engineering, as stated above, it is urgent to develop and share standardized cloning protocols specifically designed for chloroplast genome engineering, taking advantage of stable targeted insertion by homologous recombination. Here we report the development of MoCloro, an extension of the Chlamydomonas reinhardtii MoClo toolkit allowing fast combinatorial and modular assembly of complex multigenic DNA from standardized gene parts into the Chlamydomonas plastome. We also constructed a standardized destination vector that allows easy cloning of homology arms required for targeting at different plastome sites, thus increasing the versatility of this tool for chloroplast genome engineering. We expect that MoCloro facilitates the implementation of Design−Build−Test−Learn cycle pipelines to optimize multigene expression and bioproduction in microalgal cell factories.
2 MATERIALS AND METHODS
2.1 Strains and growth conditions
Wild type (WT) 137c strain of Chlamydomonas reinhardtii (CC-125 wild type mt+, Chlamydomonas Stock Center) was used. Cells were typically grown in standard Tris-Acetate-Phosphate (TAP) liquid medium under constant illumination (50 μmol m−2 s−1) at 25°C and 125 rpm in an orbital shaker Innova 42 (Eppendorf) or in incubator S600 PLH-LED (Aralab). Cell growth in liquid media was monitored by measuring the optical density (OD) at 750 nm. TAP solid medium was prepared with 1.5% agar and supplemented with spectinomycin (100 mg L−1, stocks at 100 g L−1 in water, Sigma-Aldrich Ref S4014). TAP plates were grown under constant illumination (50 μmol m−2 s−1) at 25°C.
2.2 Molecular biology protocols
PCR amplification was performed using either iProof™ High-fidelity DNA polymerase mastermix (Bio-Rad), MyTAQ™ Mix (Ecogen) or PrimeSTAR® GXL Premix Fast (Takara). Restriction enzymes and T4 DNA Ligase were obtained from New England Biolabs (NEB). For DNA fragment phosphorylations, T4 polynucleotide kinase was used (Thermo Scientific). GeneJet PCR purification and Gel extraction kits (Thermo Scientific) were employed for DNA purification. Escherichia coli DH5α strain was used to amplify plasmid DNA, which was isolated using NZY Miniprep kit (NZYtech). The list of primers used can be found in Table S1.
2.3 Biobricks design and construction
All Level 0 biobricks (Promoters-5′ untranslated regions (UTRs): rbcL, 16SrRNA, atpB, psaA and psbD; 3′UTR-Terminators: psbA-T7, atpB, psaA and srbcL; Coding Sequences: uidA) were designed and analyzed in silico using Benchling platform and according to Chlamydomonas MoClo syntax (Weber et al., 2011; Crozet et al., 2018). Gene parts were either amplified by PCR from plasmid or plastome DNA, or obtained synthetically from Twist Bioscience HQ (San Francisco, USA) or GeneArt (Thermo Fisher), as indicated in Table S2. All biobricks were cloned into the maintenance vector pTwist Chlor High Copy provided by Twist Bioscience HQ. The β-glucuronidase enzyme encoded by the uidA gene from E. coli (GUS protein) was codon harmonized using the %MinMax calculator (Rodriguez et al., 2018) according to chloroplast codon usage (available at https://www.kazusa.or.jp) and domesticated (eliminating BsaI and BbsI sites). The specific DNA sequence for each Biobrick can be found as Supplementary Data.
2.4 MoCloro Level 1 assembly reactions and generation of selection marker
Golden Gate Level 1 reactions to generate modules/Transcriptional Units (TU) from Level 0 biobricks were performed in a final volume of 20 μL, containing 20 U of BsaI-HFv2 together with 400 U of T4 DNA Ligase, 2 μL of rCutSmart buffer (NEB) and 2 μL of ATP 10 mM. DNA ratio was 100 fmol backbone (MoClo Level 1 destination vector) and 200 fmol of each insert. Reactions were run at a thermocycler with the following program: [5 min at 37°C – 5 min at 16°C]x50, 1 h at 37°C, 10 min at 60°C, 20 min at 80°C and hold at 10°C. Two μL were typically transformed into DH5α competent cells and plated and selected in LB-agar plates with ampicillin, X-Gal and IPTG.
Standard MoClo Level 1 destination vectors containing BsaI sites for TU assembly, flanked by BbsI sites for Level 2 reactions, were used and included pL1-1F, pL1-1R, pL1-2F, pL1-2R, pL1-3F and pL1-3R. These vectors allow cloning of the TU at a specific position and direction in the MoClo Level 2 destination vectors, where F refers to Forward and R refers to Reverse orientation of the TU, and the number before F or R indicates the position of the TU in the final device as described (Weber et al., 2011).
We also generated Level 1 modules containing the spectinomycin resistance (SpecR) gene as a selection marker for chloroplast transformation under the control of the atpA 5′ regulatory region and rbcL 3′ terminator. SpecR transcriptional unit was amplified from the pLM20-RhlA vector (Miró-Vinyals et al., 2023) by PCR using adapted primers that introduced BbsI sites and subcloned at BamHI and EcoRI sites of the pUC19 vector (DNA sequences are in Supplementary Data and primers in Table S1). Importantly, four different versions of the SpecR module were generated for further assembly in Level 2 vectors in forward orientation at positions one, two, three and four of multigenic devices (vectors called pCC1-23, pCC1-05, pCC1-24, pCC1-25 respectively).
2.5 MoCloro Level 2 assembly reactions
Golden Gate Level 2 reactions to generate devices/multigenic constructs from Level 1 modules/TUs were performed in a final volume of 20 μL, containing 20 U of BbsI-HF together with 400 U of T4 DNA Ligase, 2 μL of rCutSmart buffer (NEB) and 2 μL of ATP 10 mM. DNA ratio was 25 fmol backbone (MoClo Level 2 destination vector) and 50 fmol of each insert (Level 1 module). Standardized MoClo Level 2 linkers pELE-2 (for assembly of two TUs) or pELE-3 (for assembly of three TUs) were used as described (Weber et al., 2011), which allow assembly of the second or the third transcriptional unit, respectively, to the 3′ BbsI site of the Level 2 destination vector. Reactions were run at a thermocycler with the following program: [5 min at 37°C – 5 min at 16°C]x50, 1 h at 37°C, 10 min at 60°C, 20 min at 80°C and hold at 10°C. 5 μL were typically transformed into DH5α competent cells and plated and selected in LB-agar plates with kanamycin, X-Gal and IPTG. Around ten white DH5α colonies were then streaked onto two LB-agar plates, one supplemented with ampicillin and the other with kanamycin, and kanamycin-resistant and ampicillin-sensitive colonies were selected for further analysis.
2.6 Construction of MoCloro Level 2 destination vectors pWF.K.4, pK4 and pLM20.K.4
MoCloro Level 2 destination vectors for chloroplast transformation were constructed as follows. pWF.K.4 was inspired by the pWF vector (Kuras and Wollman, 1994) and constructed in two sequential steps, which are illustrated in Figure S1. First, the ampicillin resistance gene (AmpR) from pWF was replaced by a kanamycin resistance (KanR), to obtain the pWF.K vector. For this, we amplified the whole pWF vector without AmpR using PL163 and PL160 primers and the KanR from a pMK vector (GeneArt) using PL164 and PL165 primers. Both PCR fragments were purified and cloned in a manner similar to a Golden Gate Level 1 reaction (but with 100 fmol of each fragment added). Second, the LacZα cassette (amplified from pUC19 plasmid using primers PL166 & PL167 and phosphorylated using T4 polynucleotide kinase) was cloned by blunt ligation at the HincII site of pWF.K vector, obtaining the final pWF.K.4 vector.
pK4 destination vector was constructed by assembling a fragment containing the backbone sequence of pWF.K.4 vector (containing KanR and origin of replication) with the LacZα cassette from pUC19. The backbone from pWF.K.4 was amplified with PL243 and PL244 primers, whereas the LacZα cassette was amplified from the pUC19 plasmid using primers PL166 and PL167 and phosphorylated using T4 polynucleotide kinase. Purified fragments were then ligated in a blunt reaction to obtain the pK4 vector, as illustrated in Figure S4.
pLM20.K.4-3′S, and pLM20.K.4-3′L Level 2 destination vectors were obtained by cloning the pLM20 vector homology region arms into the pK4 destination vector by Golden Gate reaction. Briefly, pLM20 5′, 3′ short (3′S) and 3′ long (3′L) homology arms were amplified using primers PL237 & PL238 for 5′ arm, PL239 & PL249 for 3′S arm and PL250 & PL251 for 3′L. Purified products were cloned as 5′ and 3′ homology arms into the pK4 vector by Golden Gate reaction to obtain the pLM20.K.4-3′S vector (assembling 5′ and 3′S homology arms) and the pLM20.K.4-3′L vector (assembling 5′ and 3′L homology arms). Golden Gate reactions were performed similarly to Level 1 MoClo reactions, except that only 25 fmol of pK4 vector and 50 fmol of each homology arm were used. Colony PCRs with MyTAQ™ Mix (using primer combinations PL239 & PL249 or PL250 & PL252) respectively were performed to identify candidate clones. Constructed destination vector DNA sequences (pWF.K.4, pK4, pLM20.K.4-3′S and pLM20.K.4-3′L vectors) can be found at Supplementary Data.
2.7 Plasmid quality control
All Level 0 biobricks were confirmed by analytic digestion and Sanger sequencing (Stab Vida, Caparica, Portugal). Level 1 plasmids were confirmed by analytic digestion. Level 2 destination vectors, pK4 vector and constructed devices were confirmed by analytic digestion and sequenced by Oxford nanopore technology (Eurofins).
2.8 Chlamydomonas chloroplast transformation by particle microbombardment
Chlamydomonas chloroplast was transformed using the PDS 1000-He Particle Delivery System from Bio-Rad. 20 mL of TAP media pre-inoculums were grown for 2–3 days. Inoculums were initiated by diluting the pre-inoculum in 2 L Erlenmeyer flasks with 400 mL of TAP media to a calculated initial OD750nm of 0.0001 and grown in a 16 h light at 50 μmol m−2 s−1 and 8 h dark photoperiod at 25°C for around 3 days. When inoculum reached a cell concentration between 5·105 and 2·106 cells mL−1, cells were harvested and resuspended with TAP media to reach a concentration of 1–3·108 cells mL−1. 100 μL of concentrated cells were dispersed onto TAP-agar plates supplemented with 100 mg L−1 spectinomycin. Plates were kept in darkness until transformation. Tungsten M-10 microparticles were resuspended in 1 mL of 70% ethanol, ultrasonicated for 5 min, settled down on ice for 15 min and washed with water four times. Finally, microparticles were resuspended in 50% glycerol sterile solution to keep 3 mg of tungsten microparticles in 50 μL glycerol solution. For DNA-coating, 10 μL of minipreps (normally at a concentration between 100 ng μL−1 and 600 ng μL−1) were added together with 50 μL of CaCl2·2H2O 2.5 M and 20 μL of spermidine 0.1 M (both freshly prepared) and agitated vigorously for 10 minutes. After chilling 15 min on ice and a first wash with 250 μL of pure ethanol, DNA-coated microparticles were resuspended in 60 μL of pure ethanol. For microbombardment, 6 μL of DNA-coated M-10 tungsten microparticles were loaded on macrocarries, 1100 psi rupture disks were used, and plates were placed 6 cm away from the stopping chamber. After bombardment, plates were incubated in darkness overnight and moved to continuous light at 25°C for selection for two or three weeks.
2.9 Chlamydomonas genotyping by PCR
Homoplasmy was confirmed by PCR genotyping using the primers listed in Table S1. Total DNA from transplastomic lines was extracted as described elsewhere (Edwards, Johnstone and Thompson, 1991). Briefly, 300 μL of DNA Extraction Buffer (200 mM TRIS–HCl pH 7.5; 250 mM NaCl, 25 mM EDTA and 0.5% SDS) were used to resuspend a micropipette tip of Chlamydomonas cells taken from a TAP-agar plate. After vortex and 5 min incubation, samples were centrifuged at maximum speed (± 20.000 g) for 5 min. The supernatant was transferred to a new tube, and 300 μL of isopropanol were added, incubated 2 min and centrifuged 15 min at maximum speed (± 20.000 g). The DNA pellet was washed by resuspending in 300 μL of 70% ethanol, and pellets were dried for at least 15 min at room temperature. Finally, the DNA pellet was resuspended in 50 μL of 10 mM EDTA, and 1 μL was used for PCR amplification. PCR were done using MyTaq™ DNA Polymerase (Bioline) according to manufacturer instructions.
2.10 GUS assay
Cultures for GUS assays were grown until saturation in 2 mL deep-well plates using 1.5 mL of TAP+Spectinomycin for 3 days. OD750nm was measured, and cultures were diluted to OD750nm at 0.15 and then grown for an additional 24 h (reaching OD750nm 0.8–0.9). Cultures were pelleted by centrifugation (3000 g, 10 min). The cell pellet was resuspended in 100 μL BugBuster (Sigma Aldrich) and kept in agitation for 1 h at room temperature. Protein extracts were centrifuged at a maximum speed (more than 3000 g) for 5 min to remove cell debris.
Reactions for GUS activity were performed in 96-well plates. First, 100 μL of water was added to the plate and incubated at 37°C. 25 μL of each assayed protein extract were loaded onto the reaction plate. To start the GUS reaction, 125 μL of 2X reaction buffer (1 mM of 4-Methylumbelliferyl-β-D-glucuronide hydrate (4-MUG from Sigma Aldrich)) were added in a final reaction volume of 250 μL (final 4-MUG concentration 0.5 mM). GUS activity measurements were taken at 5, 30, 60 and 120 min after the initial addition of the reaction buffer and incubation at 37°C. Reactions were then stopped by transferring 50 μL of the reaction volume to a Stop Buffer plate containing 200 μL of 200 mM Na2CO3, and fluorescence was read at EXC 365 nm, EM 460 nm in a FLx800 plate reader (BioTek). To normalize GUS activity among samples, total protein quantification was carried out according to the protocol below, and specific GUS activity was calculated by relating the activity to the total protein present in the reaction. The wild type 137c line and a previously generated line from our laboratory containing the GUS reporter gene under the rcbL promoter and psbAT7 terminator were used as negative and positive controls, respectively. GUS activity was assayed in six independent transformants considered biological replicates. Statistical analysis was performed using GraphPad Prism software.
2.11 Protein quantification from the extracts by BCA
Protein concentration was determined by using a commercial BCA kit according to manufacturer instructions (Thermo Scientific). We used 200 μL of Reagent and 25 μL of protein extracts at a 1/4 dilution. Samples were read using Absorbance at 595 nm with an ELx808 plate reader (BioTek).
2.12 Protein extraction and immunoblot analysis of transplastomic lines
Wild-type 137c and transplastomic lines were grown in 20 mL of TAP medium to an OD750 nm of 0.25–0.35. Then, 5.25 x 107 cells were harvested by centrifugation for 10 min at 3000 g, and resuspended in 80 μL SDS-PAGE gel charge buffer (25% glycerol, 50 mM of Tris–HCl at pH 7, 1% (w/v) of SDS, 0.05% (v/v) of 2-mercaptoethanol and 0.005% (w/v) of bromophenol blue). Total protein extracts were boiled for 5 min and centrifuged at 15000 g for 5 min. The soluble fraction was recovered and separated on a 12% SDS-polyacrylamide gel electrophoresis (Bio-Rad, Hercules, USA), and transferred onto a PVDF membrane (Millipore, Burlington, MA, USA). Immunodetection was performed with a primary rat anti-HA high-affinity antibody (1:1000) (Merck) followed by a secondary mouse anti-Rat-HRP antibody (1:10000) (Merck). Chemiluminescence detection was performed using Thermo Scientific™ SuperSignal™ Chemiluminescent Substrate (ThermoScientific) and imaged using Amersham™ ImageQuant™ 800 biomolecular imager (GE Healthcare). As protein loading and transfer control, PVDF membranes were stained with Coomassie Brilliant Blue (0.1% Coomassie Blue in 10% acetic acid, 50% methanol and 40% H2O for 30 seconds; washes with 10% acetic acid, 50% methanol and 40% H2O).
3 RESULTS & DISCUSSION
3.1 Construction of standardized Level 0 biobricks and Level 2 destination vector for Chlamydomonas chloroplast transformation and transgene expression
In order to design chloroplast gene parts (biobricks) for multigene assembly, we adopted the standard syntax previously used in MoClo (Weber et al., 2011; Crozet et al., 2018). Level 0 biobricks were constructed by flanking with BsaI target sites with necessary overhangs to allow subsequent Level 1 assembly (Figure 1A). Constructed biobricks included (1) five different Chlamydomonas chloroplast 5′ regulatory regions (parts A1-B1) containing promoter and 5′ untranslated region (5′UTR): rbcL, 16SrRNA, atpB, psaA and psbD; and (2) four different 3′ regulatory regions (parts B6-C1) containing 3′UTRs and transcriptional terminators: psbA-T7, atpB, psaA and srbcL (Figure 1A and Table S2). All these regulatory sequences were previously described for chloroplast heterologous gene expression (Mcbride et al., 1994; Barnes et al., 2005; Kato et al., 2007; Michelet et al., 2011; Larrea-Alvarez and Purton, 2020). Promoter-5′UTRs and 3′UTR-terminators flanked by BsaI restriction sites were either synthetically obtained or amplified by PCR as indicated (Table S2). For the rbcL terminator, a short version of 258 bp (srbcL) was used instead of the 407 bp full length. This short version still renders functional transcriptional termination while avoiding an internal BsaI site (Larrea-Alvarez and Purton, 2020). We also generated a biobrick for expressing GUS (β-glucuronidase enzyme encoded by the uidA gene from Escherichia coli) as a reporter (parts B2-B5 in Figure 1A). We also used the hemagglutinin (HA) tag biobrick (part B5) from Chlamydomonas MoClo toolkit (pCM0-098 vector, Crozet et al., 2018) that allows C-terminal fusion with target proteins cloned between positions B2-B4 (Figure 1A). All biobricks were cloned into the maintenance vector pTwist Chlor High Copy as indicated (Table S2).
We also designed and constructed the pWF.K.4 vector, a Level 2 Golden Gate compatible destination vector, containing Chlamydomonas plastome 5′ and 3′ homology regions (arms) to allow Chlamydomonas plastome transformation by homologous recombination in the pWF locus (Figure 1B). For this, we used the pWF vector as inspiration (Kuras and Wollman, 1994), but modified it in several ways, as detailed in Figure S1. In brief, we exchanged the E. coli selection marker Ampicillin resistance (AmpR) with a Kanamycin resistance (KanR) gene and introduced the LacZ-alpha gene (coding for the alpha fragment of the β-Galactosidase) flanked by BbsI sites, resulting in the splitting of the pWF locus around the petA site of the plastome in two homology regions (pWF 5′ and pWF 3′ arms, Figure 1B and Figure S1).
DNA sequences of the constructed Level 0 biobrick and Level 2 pWF.K.4 destination vector can be found in the Supplementary Data. pWF.K.4 vector was then used as a destination vector for combinatorial functional analysis of constructed MoCloro parts.
3.2 Generation of combinatorial constructs for functional testing of biobricks
To functionally validate the generated biobricks and the pWF.K.4 vector, we constructed and transformed lines expressing the reporter GUS gene under combinations of five promoter-5′UTRs and four 3′UTR-terminators (Figure 2A). GUS transcriptional units were built for the first position and forward orientation (using standard MoClo destination vector pL1-F1), obtaining 20 different Level 1 combinations. For Level 2 reactions, the 20 different GUS TU's were combined with the module containing spectinomycin resistance (SpecR) for selection of chloroplast transformants (vector pCC1-05, see Materials&Methods and DNA sequences in Supplementary Data), and cloned into the new Level 2 destination vector pWF.K.4 (Figure 1B) for chloroplast transformation. For the generation of devices with two TUs, the pELE-2 linker vector was used in Level 2 reactions as described (Weber et al., 2011), which allows the ligation between the second TU (located downstream) and the 3′ site of the destination vector. All 20 final devices were confirmed by full plasmid nanopore sequencing. Chlamydomonas 137c cells were transformed by microbombardment, and homoplasmy was confirmed by PCR (Figure S2), after at least 5 rounds of selection in spectinomycin plates. Six independent Chlamydomonas transformant lines were randomly selected and grown in deep-well plates until the late exponential phase (OD750nm 0.8–0.9), and protein extracts from cell pellets were prepared for enzymatic GUS assay (Figure 2B). As a negative control, we used protein extracts from a wild type 137c line, whereas as a positive control, we used one line maintained in the laboratory for a long time (more than 7 subculture passages) expressing GUS under the control of rcbL promoter-5′UTR and psbAT7 3′UTR-terminator sequences (rbcL-GUS-psbAT7). This line showed stable GUS activity across independent experiments (Figure S3), thus indicating relatively low technical error (<5%) and a high reproducibility of the assay. Results showed that all tested promoter-terminator combinations are able to express detectable levels of GUS reporter (Figure 2B), thus confirming the functionality of all the constructed biobricks. Interestingly, rbcL and 16SrRNA promoter-5′UTR seem to drive higher reporter expression than that of psbD, atpB or psaA in most of the combinations tested.
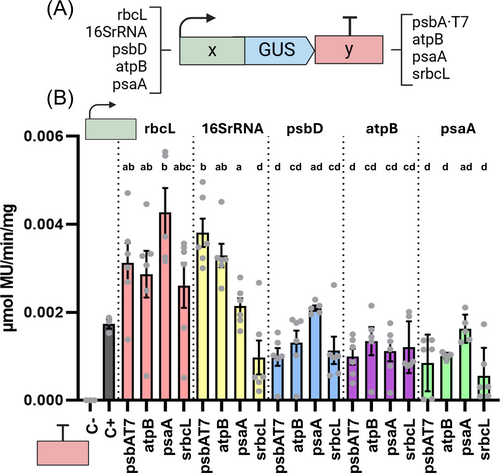
One intriguing observation is the biological variability detected between the six independent lines analyzed for each construct (Figure 2B), which ranged from low-moderate (as for the combinations with the 16SrRNA promoter-5′UTR) to moderate-high (up to 10-fold changes in extreme cases). Considering the technical confidence of the GUS assay (Figure S3), the data strongly suggests that the origin of this variation is biological. This is somehow surprising as it is generally assumed that position effects are reduced when constructs are targeted to specific plastome loci. However, we believe that biological variation of transgene expression in transplastomic lines has not been properly evaluated in Chlamydomonas. Instead, studies tend to show the results of a relatively small number of selected lines (i.e. not random) that behave similarly. The source of this biological variability is unknown, although we can speculate on a number of factors, such as the different physiological status of the cell or the number of plastome copies in the chloroplast. Noteworthy, the observed variation is in agreement with previous results from our laboratory, showing more than 3-fold change differences in product accumulation (Miró-Vinyals et al., 2023).
Because of the relatively high biological variation, it was difficult to assess the effect of the 3′UTR-terminator in the reporter GUS expression for each Promoter-5′UTR combination, as differences were not statistically significant (Figure 2B). In the case of the 16SrRNA promoter-5′UTR, however, the 3′UTR-terminator establishes significant differences in reporter GUS expression, from the highest (psbA-T7 3′UTR-terminator) to the lowest (srcbL 3′UTR-terminator). Together, these results are in agreement with other studies indicating that promoter-terminator combinations drive significant variation in gene expression (Barnes et al., 2005; Inckemann et al., 2024), and reinforce the importance of performing combinatorial screening and analysis of several independent transformant colonies to identify those constructs that express desired levels of expression of the gene of interest. Interestingly, recent analysis indicates that the 5′UTR region is especially relevant for driving high levels of transgene expression (Inckemann et al., 2024). By constructing new biobricks, MoCloro can be easily adapted to generate chimeric promoters and 5′UTR constructs.
3.3 Testing multigene construction and expression
To further characterize the functionality of MoCloro for assessing complex designs, we built a set of multigene combinations aimed at testing gene orientation and position effect in the context of 3 gene expression devices (Bock, 2013). For this, we constructed several Level 1 modules including (1) the GUS reporter gene under the control of rbcL and psbA-T7 as 5′ and 3′ regulatory sequences, respectively (rbcL-GUS-psbA-T7); (2) the SpecR cassette (atpA-SpecR-rbcL); and (3) the gene encoding RhlA acyltransferase from Pseudomonas aeruginosa fused to an HA tag under the control of 16SrRNA and atpB as 5′ and 3′ regulatory sequences, respectively (16SrRNA-RhlA-HA-atpB). This gene was selected as proof of the principle of our MoCloro multigene design, as it was previously expressed in Chlamydomonas chloroplast by conventional cloning in the pLM20 vector (Miró-Vinyals et al., 2023). We generated a new biobrick by cloning the codon-optimized RhlA coding sequence at positions B2-B4 (Table S2), allowing the fusion to the HA tag (B5 site) according to the MoCloro design (Figure 1A). From Level 1 modules, we generated six different devices containing GUS reporter cassettes at first, second and third position, in forward (1F, 2F, 3F, using the MoClo destination Level 1 vectors pL1-1F, pL1-2F or pL1-3F respectively) or reverse (1R, 2R, 3R, using the MoClo destination Level 1 vectors pL1-1R, pL1-2R or pL1-3R respectively) orientation. In these devices, SpecR and RhlA cassettes were located in forward orientation, at positions one and two for SpecR (from MoClo destination Level 1 vectors pL1-1F and pL1-2F) or at positions one and three for RhlA (from the MoClo destination Level 1 vectors pL1-1F and pL1-3F). The three Level 1 modules (GUS, SpecR, RhlA, combined with the required pELE-3 linker) were assembled using our generated Level 2 destination vector pWF.K.4 (Figure 3A).
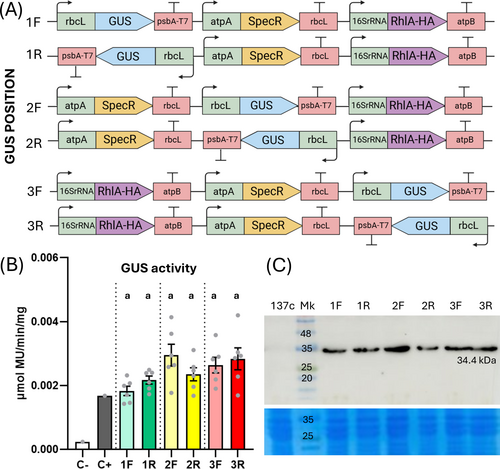
Upon 137c strain transformation with the multigenic construct devices, four rounds of selection were performed in spectinomycin plates. Six independent transplastomic lines for each construct were assayed for GUS activity. This analysis revealed no significant differences among tested combinations (Figure 3B). Importantly, expression of the other gene (RhlA-HA) in the devices was detected in all constructs by Western blot (Figure 3C), thus indicating the feasibility of expression of at least two genes of interest in our multigenic assembly design.
A recent manuscript suggests that termination of transcription might not be well defined by some Chlamydomonas terminators, such as the one from the psbH gene (Navarrete and Pollak, 2023), so that antisense transcription of proximal genes located in reverse orientation might trigger gene silencing. We assayed some constructs containing the SpecR and the GUS cassettes in antisense orientation (2R and 3R, Figure 3A). We did not observe a significant effect of these constructs on the spectinomycin-resistant phenotype or the GUS expression level (Figure 3B), thus indicating that the used atpA/rbcL and the rsbcL/psbA-T7 promoter/terminator combinations of SpecR and GUS cassettes, respectively, do not induce antisense silencing due to poor termination. This effect might be dependent on the strength of the promoter combined with the capacity to stop transcription of the terminator. The versatility of MoCloro allows easy testing of new multigenic combinations.
3.4 Construction of a new vector to enable new integration sites in Chlamydomonas plastome
A great advantage of chloroplast engineering is that transgenes can be easily integrated at different plastome sites in a targeted manner by replacing the flanking 5′ and 3′ homology arms. This possibility allows the optimization of transgene expression by identifying favourable plastome contexts and facilitates complex designs requiring multiple integration sites. The constructed standardized destination vector pWF.K.4 proved to be functional for plastome transformation and transgene expression at the pWF locus (Figures 1 and 2). We next aimed to increase the versatility of the developed MoCloro by designing and constructing the vector pK4, a novel standardized Level 2 destination vector that facilitates Golden Gate-based cloning of new 5′ and 3′ homology arms, thus enabling integration at desired plastome loci. pK4 was constructed as described in Figure S4 and is represented in Figure 4A. pK4 DNA sequence is provided as Supplementary Data. Similar to pWF.K.4, the pK4 vector contains the Kanamycin resistance gene (KanR) as an E. coli selection marker and the LacZ-alpha gene flanked by MoClo-compatible BbsI sites. However, instead of the pWF 5′ and 3′ arms, the pK4 vector contains two BsaI sites strategically located at 5′ and 3′ ends of the BsbI-LacZα-BsbI fragment. The BsaI sites are used to introduce the user-chosen plastome 5′ and 3′ homology regions previously amplified by PCR in a one-pot Golden Gate assembly reaction. Once homology regions are introduced, BsaI sites disappear, and the obtained novel p”XX”K4 vector can be directly used as Level 2 MoClo destination vector for chloroplast transformation (Figure 4A).
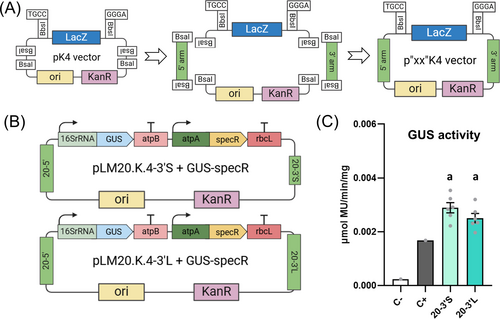
To validate pK4 functionality, we constructed two different Level 2 destination vectors using different versions of 5′ and 3′ homology arms around the chloroplast genomic loci psbA and 23S, which were amplified by PCR from pLM20 vector (Miró-Vinyals et al., 2023). As 5′ arm (20–5′), we used the 1.7 kb long homology region from pLM20. To avoid BbsI sites located in the middle of the 3′ region of the pLM20 homology arm, we employed two different, non-overlapping, 3′ arms: a short fragment upstream of BbsI sites (0.6 kb, 20-3′S) and a downstream long fragment (1.3 kb, 20-3′L). Both short and long 3′ arms should result in a very similar integration site in the plastome. Assembly reactions of these fragments with the pK4 vector resulted in the construction of Level 2 destination vectors pLM20.K.4-3′S and pLM20.K.4-3′L vectors as detailed in Material & Methods. After validation of the vectors by full plasmid nanopore sequencing, both plasmids were employed as destination vectors in Level 2 Golden Gate reactions to assemble a GUS transcriptional unit (16SrRNA-GUS-atpB) together with the SpecR selectable marker to generate pLM20.K.4-3′S + GUS-SpecR and pLM20.K.4-3′L + GUS-SpecR (Figure 4B). After Chlamydomonas chloroplast transformation by microbombardment, both plasmids generated transplastomic lines resistant to spectinomycin, thus confirming the functionality of the used 20 5′ arm and 20 3′ short and long homology arms.
As expected, the pLM20.K.4 integration site was also suitable for heterologous gene expression, and GUS activity was detected in six independent transformant lines (Figure 4C). Moreover, the mean specific GUS activity was similar between these lines (Figure 4C) and the lines that integrated an equivalent 16SrRNA-GUS-atpB transcriptional unit at the pWF integration site (Figure 2B), suggesting a minor effect of the integration site in the expression levels of this GUS cassette.
Both short and long 20–3′ arm versions of constructed pLM20.K.4 vectors (Figure 4B) rendered transformant colonies, which is consistent with previous data demonstrating that homologous recombination events can happen with DNA fragments shorter than 600 bp (Dauvillee et al., 2004). However, we noticed that the long 20–3′ arm version (1.3 kb) in pLM20.K.4-3′L vector showed higher transformation efficiency than the short one (0.6 kb) (Table S3), indicating that longer arms favour homologous recombination. Interestingly, the homology arms of pLM20 recombine at the inverted repeat region of the plastome. Hence, once homoplasmy is reached, the number of transgenes potentially doubles compared to integration sites that are unique, such as the pWF site. However, when both integration sites (pWF vs. pLM20) were compared (Figure 2B vs. 4C) the reporter GUS activities proved to be very similar, suggesting that transgene expression levels are mostly independent of plastome context, in agreement with other authors (Inckemann et al., 2024). Together, the developed pK4 vector enables the generation of new integration sites in the plastome in an easy and fast manner, thus expediting Chlamydomonas chloroplast engineering.
4 CONCLUSIONS
To produce and optimize bioproducts of interest in microalgae, it is necessary to establish efficient protocols for vector construction, transformation, line characterization and selection of the best performers. This process is facilitated by combinatorial designs and high-throughput screening methodologies according to the Design-Build-Test-Learn cycle pipeline. Transgene expression often requires optimization by testing different combinations of regulatory elements and/or coding sequences. Moreover, bioproduction might also require expressing multiple genes in complex multigenic constructs. Here, we established a methodology to facilitate vector construction and multigene assembly for Chlamydomonas plastome transformation and transgene expression (MoCloro). The design was based on the Chlamydomonas MoClo toolkit (Crozet et al., 2018) established for nuclear transformation and expression. In this work, we constructed five promoter-5′UTRs, four 3′UTR-terminators and one reporter (GUS, Figure 1), and confirmed their functionality by combinatorial analysis (Figures 2, 3 and 4). Our library of pieces can be further expanded by cloning novel promoters, 5′UTRs, reporters, tags and terminators following MoClo syntax. The tool can also be adapted to incorporate recent advances that facilitate a more direct cloning from Level 0 to Level 2 (Niemeyer and Schroda, 2022). Importantly, we developed a vector (pK4) that facilitates cloning of 5′ and 3′ homology arms for the design of customized integration site. In our hands, we managed to construct final Level 2 vectors in as short as 10 days, starting from Level 0 biobricks (Figure 5). Upon vector confirmation by Oxford nanopore technology sequencing, transplastomic lines are selected and restreaked until homoplasmy is achieved. The whole process from Level 0 biobricks to line characterization might be completed in less than two months (Figure 5), or even faster if homoplasmy is not required. This work represents a significant advancement facilitating more efficient and rapid cloning for chloroplast engineering in green microalgae, expediting the process of generation, screening and selection of the best-expressing constructs.
AUTHOR CONTRIBUTIONS
PL and AP, conceptualization. XM-C and MG designed, performed, analyzed the experiments and wrote the original manuscript. AP and EM acquired funds. AP, PL, MC and EM, supervised and discussed data. All authors contributed to the final manuscript.
ACKNOWLEDGEMENTS
pWF was kindly provided by Katia Wostrikof (IBPC, Paris). We acknowledge Jae-Seong Yang (CRAG, Barcelona) for kindly providing materials. We thank Montserrat Capellades (CRAG, Barcelona) for her technical support implementing microbombardment system. We thank Sofia Campos and Marc Mialet (Laboratory of Biochemistry, IQS-URL) for their technical support in biobrick construction. This work was supported by GLYCOENGIN (PID2022-138252OB-I00) to A.P. and RETROPhot (PID2021-122288NB-I00) to E.M. both funded by MICIU/AEI/10.13039/501100011033 and by ERDF/EU, and by Projects 2021 SGR-00535 to A.P. and 2021 SGR-00792 to E.M., both funded by AGAUR agency/Generalitat de Catalunya. E.M. acknowledges financial support from the Spanish Ministry of Economy and Competitiveness, through the ‘Severo Ochoa Programme for Centres of Excellence in R&D' CEX2019-000902-S funded by MICIU/AEI/10.13039/501100011033 and the CERCA Programme/Generalitat de Catalunya. M.G. acknowledges a Beatriu de Pinòs postdoctoral fellowship from AGAUR agency/Generalitat de Catalunya and XM-C acknowledges a predoctoral fellowship from Institut Químic de Sarrià.
Open Research
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the supplementary material of this article. Biobrick sequences and plasmids constructed in this work are deposited at the Chlamydomonas Resource Center (https://www.chlamycollection.org/). Other materials are available from the corresponding authors upon reasonable request.




