PbRVE6 Promotes Anthocyanins Accumulation in Pear Fruit Peel by Regulating Key Biosynthetic Genes
Abstract
Red peel color, a desirable trait in pears, is determined by anthocyanin accumulation. While REVEILLE (RVEs) transcription factors regulate anthocyanin biosynthesis in some plant species, their role in pear peel has not been well-studied. This study investigates the function of RVEs in anthocyanin biosynthesis in ‘Zaosu’ (low anthocyanin content) and its red bud mutant, ‘Red Zaosu’ (high anthocyanin content) fruit peel. Consistent with higher anthocyanin content, ‘Red Zaosu’ pears exhibited increased PbRVE6 expression compared to ‘Zaosu’, while PbRVE3a and PbRVE3b levels remained unchanged. Additionally, PbRVE6 was localized to the nucleus. Overexpression of PbRVE6 in ‘Zaosu’ pear pericarp significantly increased anthocyanin content and upregulated key anthocyanin pathway genes PbANS and PbUFGT. Conversely, VIGS silencing of PbRVE6 in ‘Red Palacer’ pears led to decreased expression of PbANS and PbUFGT and a slight reduction in anthocyanin content. Yeast one-hybrid and dual-luciferase assays confirmed that PbRVE6 can bind and activate the promoters of PbANS and PbUFGT. These findings demonstrate that PbRVE6 promotes anthocyanin accumulation in pear peel by directly regulating PbANS and PbUFGT expression. This study provides a valuable foundation for understanding the regulatory network of anthocyanins in pear peels, and offers potential genetic resources for the production of new pear germplasm.
1 INTRODUCTION
Anthocyanins are key secondary metabolites found widely in fruits and vegetables, contributing to the vibrant color of red pears and offering potential health benefits for humans (Wedick et al., 2012; Kaur et al., 2023). Red-skinned pears, with their superior appearance and possible health benefits, represent a valuable germplasm resource with solid market competitiveness (Liu et al., 2023a). Additionally, anthocyanins play a crucial role in regulating plant growth and development and mitigating oxidative damage caused by environmental stressors such as ultraviolet radiation and infection (Chachar et al., 2024). Anthocyanin biosynthesis in plants is stimulated by multiple factors, including intracellular signals and exogenous environmental stressors (Honda & Moriya, 2018; Liu et al., 2018). Understanding the regulation of anthocyanin biosynthesis in pears could improve fruit quality and potentially higher economic gains for pear growers.
In plants, late biosynthesis genes (LBGs) and early biosynthesis genes (EBGs) are key genes involved in the biosynthesis of anthocyanins. LBGs (including dihydroflavonol-4-reductase, anthocyanin synthase, and UDP-glucose: flavonoid-3-O-glucosyltransferase) are regulated by MYB-bHLH-WD40 complexes, while EBGs (including chalcone synthase, chalcone isomerase, and flavonoid-3′-hydroxylase) are regulated by independent R2R3-MYB transcription factors (TFs; Xu et al., 2015; Li et al., 2020).
A family of MYB-like TFs forms part of the core circadian clock and is highly conserved in land plants. The CCA1 and LHY genes belong to a larger clade of MYB TFs, including eight REVEILLE (RVE) genes, most exhibiting circadian-regulated expressions (Ahn et al., 2021). Growing evidence suggests that RVE proteins play significant roles in various biological processes. For instance, mutations in AtRVE4, AtRVE6, and AtRVE8 in Arabidopsis thaliana result in a longer circadian period, larger cells, and delayed flowering (Farinas & Mas, 2011; Scandola et al., 2022; Hughes et al., 2024). AtRVE1 regulates auxin biosynthesis genes and promotes free auxin production during the day in Arabidopsis thaliana (Rawat et al., 2009). Ectopic overexpression of SgRVE6 enhances cold tolerance in tobacco (Chen et al., 2020), and GmRVE8a appears to positively regulate responses to salt and drought stress in soybean, improving tolerance when overexpressed in Arabidopsis thaliana (Bao et al., 2024). In Arabidopsis thaliana, AtRVE8 activates the promoter of anthocyanin structural genes, regulating circadian anthocyanin accumulation (Pérez-García 2015). Additionally, the length of AT simple repeats in the 3′ untranslated region of the BoRVE5 gene is inversely correlated with anthocyanin content (Ahn et al., 2021). Studies in white pear have shown that PbRVE family genes exhibit a typical circadian pattern, peaking before and after dawn (Li et al., 2020; Liu et al., 2023b). These findings suggest that RVEs, as key regulators of the circadian system, play a significant role in the anthocyanin biosynthetic pathway. However, research on the function of PbRVE family members in red pear anthocyanin regulation remains limited.
This study aims to understand the regulation of anthocyanin biosynthesis by PbRVE TFs in pears. We investigated the expression patterns of PbRVE genes and compared them between the red bud mutant, ‘Red Zaosu’, and its wild type cultivar, ‘Zaosu’. We further explored the functional role of PbRVE6 through transient overexpression and VIGS analyses, demonstrating its positive regulation of anthocyanin biosynthesis. Our findings expand the understanding of the regulatory network governing pear anthocyanin accumulation and provide valuable gene resources for enhancing fruit appearance quality.
2 MATERIALS AND METHODS
2.1 Experimental plant materials
Fruits of Pyrus bretschneideri L. cultivars ‘Zaosu’ and red bud mutant ‘Red Zaosu’ were harvested 45 days after full bloom. The samples were rapidly frozen in liquid nitrogen and ground into powder for subsequent analysis of anthocyanin content and total RNA extraction.
For transient experiments, young fruitlets were grown on pear trees and not harvested during the experiment. The ‘Zaosu’ and ‘Red Palacer’ (P. communis L.) were used as materials for transient overexpressing and virus-induced gene silencing (VIGS) experiments of the PbRVE6 gene, respectively. To remove the red color background, ‘Red Palacer’ fruits were bagged until the skin turned white. These white fruits were then chosen for VIGS transient injections. After injection procedures, the pear fruits were exposed to natural light and harvested on the fifth day. Three independent biological replicates were collected for each experiment. After harvesting, all samples were immediately frozen in liquid nitrogen and stored at −80°C.
2.2 Determination of anthocyanin content in pear peels
Anthocyanin content in peels of pears was determined using a method described by Li et al., (2021) with minor modifications. Briefly, 0.02 g of peel powder was mixed with 1.5 mL of 1% (v/v) hydrochloric acid-methanol solution in a pre-cooled mortar. The supernatant was collected after centrifugation at 4°C and 12 000 g for 5 min. Two hundred microliters (200 μL) of the supernatant were added to 800 μL of potassium chloride buffer (250 mM, pH 1.0) and 800 μL of sodium acetate buffer (400 mM, pH 4.5). The solutions were mixed and incubated in the dark for 15 minutes. Absorbance values were measured at 520 nm and 700 nm using a spectrophotometer (Multiskan GO, Thermo Scientific). Total anthocyanin content was expressed as mg g−1 fresh weight.
2.3 Sequence analysis of PbRVE3a, PbRVE3b and PbRVE6
Total RNA was extracted from the peels using the RNAprep Pure Plant RNA Kit (Tiangen). One μg of total RNA was reverse transcribed into cDNA using the PrimeScript RT reagent kit with gDNA Eraser (TaKaRa). Primers for fluorescence quantitative PCR (qRT-PCR) were designed using the Oligo 7 software based on the coding sequence (CDS) of PbRVEs. Primers with high specificity were selected and synthesized. The NCBI accession numbers are listed in Supplementary Table S1; primers are provided in Supplementary Table S2. The qRT-PCR was performed using the TB Green Premix Ex Taq II (Tli RNaseH Plus; TaKaRa) on an Applied Biosystems StepOnePlus™ Real-Time PCR system (Applied Biosystems). The 2-ΔΔCT method was utilized to estimate the relative transcription values for RT-qPCR normalization, using PbActin7 (LOC125473976) as the reference gene.
2.4 Subcellular localization analysis of PbRVE6 in tobacco leaves
The CDS of the PbRVE6 gene was cloned into the pCambia2300-GFP vector, resulting in the plasmid 2300-PbRVE6. The construction primers for the vector are provided in Table S2. Agrobacterium tumefaciens strain EHA105 containing the constructed 2300-PbRVE6 plasmid was infiltrated into 4-week-old tobacco leaves. After infiltration, the tobacco leaves were incubated for three days in a growth chamber with a 16/8 hours light/dark photoperiod at 22°C. Fluorescence in the transformed cells was detected using an automated microscopy system (BX63, Olympus).
2.5 Transient transformation the PbRVE6 gene in young pear fruits
We used a transient transformation system to verify the gene function of PbRVE6, with minor modifications according to the method used by Li et al., (2021). The CDS of the PbRVE6 gene was cloned into the pGreenII 0029 62-SK vector using the BamHI and HindIII restriction enzyme sites, resulting in the overexpression vector PbRVE6-OE (see Table S2 for the primers used in vector construction). A. tumefaciens strain EHA105 containing the constructed plasmids was grown at 28°C on Luria-Bertani (LB) solid medium with 50 mg l−1 kanamycin and 25 mg l−1 rifampicin. After 48 h, the strain was resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES, and 200 μM acetosyringone) and shaken for 3–4 h to a final OD600 of 0.8 before injection into pear fruitlet skins. For injection, 10 bagged ‘Zaosu’ fruits were injected with the empty vector (pGreenII 0029 62-SK), and 10 bagged ‘Zaosu’ fruits were injected with PbRVE6-OE. VIGS assays were used to silence PbRVE6 in ‘Red Palacer’ pear fruits. The 421 bp fragment of the C-termini of RVE6 was inserted into the MCS (BamHI-XhoI) of pTRV2 to form PbRVE6-TRV vectors. For injection, ten bagged ‘Red Palacer’ fruits were injected with the empty vector (pTRV1: pTRV2 = 1: 1, v/v), and ten bagged ‘Red Palacer’ fruits were injected with PbRVE6-TRV (pTRV1: PbRVE6-TRV = 1: 1, v/v). After five days of light treatment, injected fruitlets were harvested for RNA extraction and anthocyanin content determination.
2.6 Yeast one-hybrid of the PbRVE6 gene
To verify the regulation of the PbRVE6 gene on its downstream genes, we used the yeast one-hybrid (Y1H) experiment referring to Li et al., (2021) with minor modification. Promoter regions of PbDFR, PbANS, and PbUFGT genes were amplified and cloned into the pAbAi plasmid using restriction enzymes (details in Supplementary Table S2). The entire PbRVE6 coding sequence was PCR-amplified and inserted into the pGADT7 vector to create the AD-PbRVE6 vector (Figure S2A). The pAbAi constructs containing promoters were linearized, and both these and the AD-PbRVE6 vector were transformed into Y1HGold yeast cells. Transformed yeast cells were plated on special media lacking leucine but containing Aureobasidin A (AbA) to select for interactions between the PbRVE6 protein and the promoter regions.
2.7 Dual Luciferase analysis of the PbRVE6 gene in tobacco leaves
To investigate the regulation of downstream genes by the PbRVE6 gene, promoters of PbDFR, PbANS, and PbUFGT were cloned into the pGreenII 0800-LUC vector (contains a luciferase reporter gene) using HindIII and BamHI restriction sites (Supplementary Table S2 for primer details). The full coding sequence of PbRVE6 was cloned into the pGreenII 0029 62-SK vector using BamHI and HindIII restriction sites (Figure S2A). A. tumefaciens containing the recombinant plasmid with PbRVE6 was infiltrated into four-week-old tobacco leaves. After infiltration, a dual-luciferase assay (dual-LUC) was performed to measure both LUC activity driven by the promoters and Renilla luciferase activity used for normalization. The ratio of LUC to Renilla luciferase activity was calculated to assess the regulatory effects of PbRVE6 on the promoters.
2.8 Data analysis
Statistical analyses were performed using the GraphPad Prism 7.0 software (GraphPad Prism). Two-tailed, unpaired Student's t-tests were employed to assess significant differences between experimental groups. Significance levels are indicated by asterisks: *: p < 0.05, **: p < 0.01. Data are presented as means ± standard errors (SEs).
3 RESULTS
3.1 Screening and identification of the PbRVE6 transcription factor
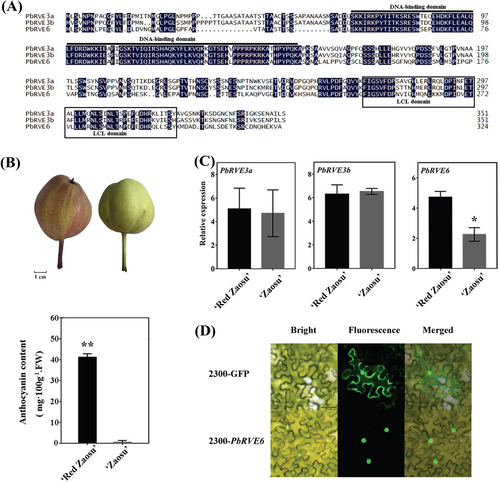
To identify candidate PbRVEs TFs involved in anthocyanin accumulation in ‘Red Zaosu’ pear fruit peels, we analyzed the amino acid sequences of PbRVE3a, PbRVE3b, and PbRVE6. Multiple sequence alignments revealed that all three PbRVEs share conserved domains: a SHAQKYF motif in their N-terminal DNA-binding domains and an LHY/CCA1-LIKE (LCL) domain in the C-terminal regions (Figure 1A). Phylogenetic analysis also clustered PbRVE6 with AtRVE6 (Figure S1). The ‘Red Zaosu’ pear, a red bud transformation of ‘Zaosu’, has red peel and leaves and exhibits higher anthocyanin content compared to ‘Zaosu’ pears (Figure 1B). Notably, the expression levels of PbRVE3a and PbRVE3b did not differ between the ‘Red Zaosu’ peel and ‘Zaosu’ pears (Figure 1C). However, the expression level of PbRVE6 in ‘Red Zaosu’ was significantly higher than in ‘Zaosu’ (Figure 1C). Subcellular localization experiments using 2300-PbRVE6 revealed a fluorescent signal in the nucleus, indicating that PbRVE6 functions as a nuclear protein (Figure 1D). These findings suggest a potential link between PbRVE6 and anthocyanin accumulation.
3.2 Functional analysis of the PbRVE6 gene
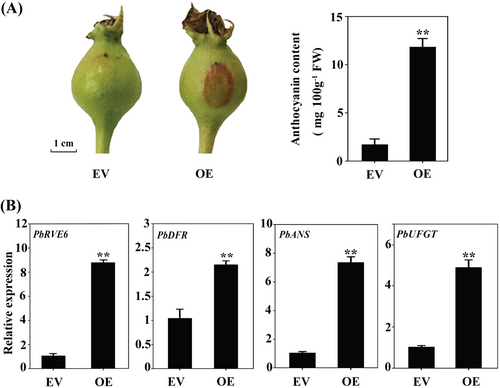
We investigated the potential role of PbRVE6 through a transient expression assay in pear fruits. Overexpression (OE) of PbRVE6 significantly enhanced pigment accumulation in the ‘Zaosu’ peels, resulting in increased red coloration at the injection site and a six-fold increase in anthocyanin content (Figure 2A). qPCR results demonstrated that PbRVE6 overexpression significantly upregulated the expression of PbDFR, PbANS, and PbUFGT genes involved in anthocyanin biosynthesis (Figure 2B). These results indicate that PbRVE6 positively regulates anthocyanin accumulation in pears.
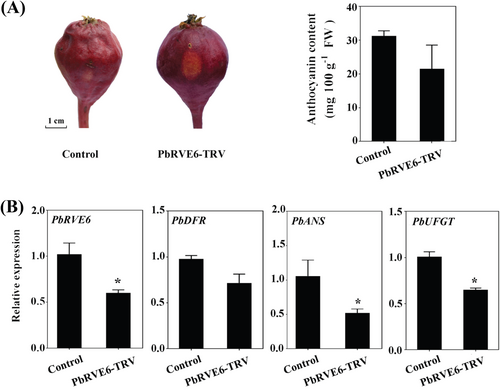
We adopted a VIGS approach, targeting the non-conserved C-terminal domain of the PbRVE6 protein sequence (Figure 1A). Silencing the PbRVE6 gene resulted in only light red coloration at the injection site (Figure 3A). Spectrophotometric measurements showed that anthocyanin content in PbRVE6-TRV fruits was reduced by one-third compared to control fruits injected with the empty vector (Figure 3B). qPCR indicated that while the expression of PbDFR remained unchanged in PbRVE6-TRV fruits, the expression levels of PbANS and PbUFGT genes were reduced (Figure 3B). These findings suggest that PbRVE6 functions as a transcriptional activator, modulating the expression of structural genes in the anthocyanin biosynthetic pathway and promoting anthocyanin synthesis in the ‘Red Zaosu’ pear fruit peels.
3.3 Regulatory effect of PbRVE6 on downstream genes of the anthocyanin biosynthesis pathway
To verify the regulatory role of PbRVE6 on the promoter regions of PbDFR, PbANS, and PbUFGT genes, we employed a Y1H experiment and a dual-LUC reporter system. Promoter regions of PbDFR, PbANS, and PbUFGT were cloned into a pAbAi plasmid to generate bait constructs for the Y1H assay (Figure S2A). The results revealed that PbRVE6 can directly bind to the promoter regions of PbANS and PbUFGT but not PbDFR, suggesting its potential role in regulating their expression (Figure 4A).
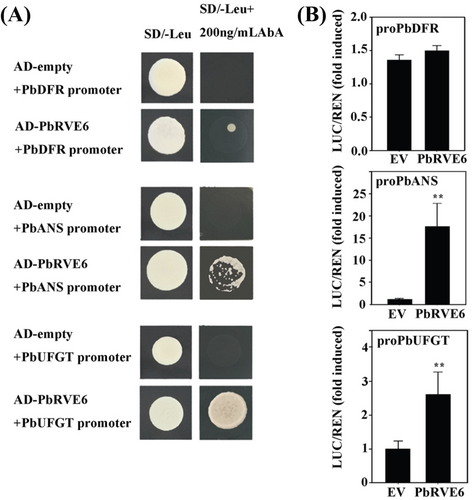
Further, dual-LUC reporter tests in tobacco leaves were conducted to verify the transcriptional activation function. Promoter fragments of PbDFR, PbANS, and PbUFGT were fused with the LUC gene to generate a reporter vector, and pGreenII 62-SK, an overexpression vector of PbRVE6, was used as the effector. An empty vector served as a negative control (Figure S2B). When the reporter construct was co-transfected with the PbRVE6 plasmid, PbRVE6 activated the reporter gene by acting on the promoters of PbANS and PbUFGT (Figure 4B). However, no activation signal was detected in the PbDFR reporter gene (Figure 4B). This activation effect provides strong evidence that PbRVE6 directly regulates the expression of these genes in pears.
4 DISCUSSION
The REVEILLE (RVE) family is a subgroup within the MYB superfamily of TFs (Farinas et al., 2011; Liu et al., 2023b). In Arabidopsis thaliana, AtRVE proteins are categorized into two subfamilies based on the presence of MYB and LCL domains. The LCL subfamily proteins, which includes AtRVE3, 4, 5, 6 and 8, features a conserved LCL domain in addition to the MYB domain (Farinas et al., 2011). Our sequence alignment analysis indicated that the PbRVE6 protein shares similar sequence features with the LCL subfamily in Arabidopsis thaliana, including a SHAQKYF-class MYB domain and an LCL domain. Phylogenetic analysis further showed that PbRVE6 clusters with the LCL subfamily proteins, particularly AtRVE6 (Figure S1). The LCL domain, exclusive to the RVE/LCL subclass, interacts with NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED proteins (LNKs), while the MYB domain provides DNA-binding specificity (Ma et al., 2018). Previous studies have shown that RVEs and LNKs function as co-transcriptional activators that promote circadian rhythm gene expression but exhibit antagonistic effects in regulating anthocyanin accumulation (Pérez-García et al., 2015). Given that PbRVE3a, PbRVE3b, and PbRVE6 possess the SHAQKYF-class MYB and LCL domains (Figure 1A), we hypothesize that PbRVE6 may interact with other TFs.
RVE genes regulate diverse processes, including the circadian clock, flowering, anthocyanin biosynthesis, hormone signaling, and stress responses in Arabidopsis thaliana (Rawat et al., 2009; Li et al., 2019). Notably, AtRVE8/LCL5 in Arabidopsis thaliana directly regulates anthocyanin gene expression in response to light (Pérez-García et al., 2015). Similarly, BoRVE5b in cabbage correlates with anthocyanin and BoDFR expression (Ahn et al., 2021). These studies suggest that RVE genes regulate anthocyanin biosynthesis across various plant species. Our study focused on three PbRVE genes differentially expressed in pear fruits. We observed that PbRVE6 expression correlated with anthocyanin content in the peels of ‘Red Zaosu’ pears compared to ‘Zaosu’ pears (Figure 1B-D), highlighting the potential of PbRVE6 to enhance the visual appeal and market value of red pears.
Anthocyanin biosynthesis relies on key enzymes such as PAL, F3H, ANS, and UFGT (Heppel et al., 2013; Honda & Moriya, 2018). The ANS-catalyzed conversion of leucoanthocyanidins to colored anthocyanidins is crucial, with ANS gene expression positively correlated with anthocyanin accumulation (Xie et al., 2003; Zhou et al., 2010). Similarly, VvUFGT expression is associated with anthocyanin production in colored grape skins (Kobayashi et al., 2001). In Crocus sativus, a MYB-related REVEILLE-8 type TF, CstMYB1R1, upregulates anthocyanin biosynthesis genes to enhance anthocyanin levels by binding to the promoter of the CstLDOX gene (Bhat et al., 2023). AtRVE8 up-regulates anthocyanin gene expression in Arabidopsis thaliana by directly binding to their promoters (Pérez-García et al., 2015). Our findings demonstrate that PbRVE6 upregulates anthocyanin content in pear fruit peels. Quantitative measurements confirmed that overexpression of PbRVE6 in ‘Zaosu’ significantly increased anthocyanin accumulation and LBG expression levels (Figure 2). Conversely, silencing PbRVE6 decreased anthocyanin content and the expression levels of PbANS and PbUFGT in pears (Figure 3). Furthermore, PbRVE6 directly binds to the promoters of PbANS and PbUFGT, activating their expression (Figure 4). These results suggest that PbRVE6 functions as a positive regulator of anthocyanin biosynthesis in pear peels by upregulating PbANS and PbUFGT gene expression. However, silencing PbRVE6 did not completely inhibit anthocyanin synthesis, suggesting the involvement of other TFs.
Given the conserved DNA-binding and LCL domains of PbRVE6, we speculate that PbRVE6 could interact with PbLNK proteins in regulating anthocyanin accumulation in pears. Xie et al., (2014) indicated that AtLNK1 and AtLNK2 regulate AtPRR5 expression by interacting with AtRVE4 and AtRVE8 to form complexes. The MYB-like TF AtRVE8 interacts with AtLNK1 and AtLNK2 to promote the expression of evening-phased clock genes and cold tolerance factors (Sorkin et al., 2023). Moreover, AtLNK proteins can prevent AtRVE8 from binding to the anthocyanin gene promoters in Arabidopsis thaliana (Pérez-García et al., 2015). Based on these findings, we proposed a model elucidating the molecular mechanism through which PbRVE6 regulates anthocyanin biosynthesis in pears (Figure S3). The model shows that PbRVE6 can promote anthocyanin accumulation by regulating the promoter activity of PbANS and PbUFGT. In addition, PbRVE6 may interact with PbLNK1, PbLNK2, PbLNK3, or PbLNK4 proteins to regulate anthocyanin accumulation in pear peels. Future studies should focus on these PbLNK TFs to provide further insights into the regulatory network governing pear anthocyanin biosynthesis and offer valuable avenues for future research.
AUTHOR CONTRIBUTIONS
X.L. wrote the original draft. X.L., W.D., S.B.H. and Z.L. reviewed and edited the manuscript. The validation was done by X.L. and W.D. The project administration was the responsibility of X.L. and W.D. The investigation was done by X.L., W.D. and X.D. The data were curated by X.L., W.D. and Y.C. The methodology was performed by L.Y., L.X. and Z.L. For the software X.D. and S.B.H. were responsible. Supervision was done by Z.L. and T.W. The formal analysis was performed by L.X. and funding acquisition was on X.L., W.D. and T.W. All authors have read and agreed to the published version of the manuscript.
ACKNOWLEDGMENTS
The authors extend our gratitude to Prof. Lingfei Xu (College of Horticulture at Northwest A&F University) for providing valuable materials, and we thank Dr. Syed Bilal Hussain (University College Dublin) for polishing the manuscript and providing valuable comments and suggestions.
FUNDING INFORMATION
This research was funded by the National Natural Science Foundation of China (32402515, 32202420) and the Youth Science Foundation of Hubei Academy of Agricultural Sciences (2023NKYJJ20, 2023NKYJJ19).
CONFLICT OF INTEREST STATEMENT
All authors declare no known competing financial interests.
Open Research
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the supplementary material of this article. Further inquiries can be directed to the corresponding author upon reasonable request.




