Loss of Arabidopsis matrix metalloproteinase-5 affects root development and root bacterial communities during drought stress
Funding information: Kempestiftelserna; Vetenskapsrådet, Grant/Award Number: 2019-04472; UC Berkeley; Umeå Universitet; Swedish Research Council
Abstract
Matrix metalloproteinases (MMPs) are zinc-dependent endo-peptidases that in mammals are known to be involved in remodeling the extracellular matrix (ECM) in developmental and pathological processes. In this study, we report At5-MMP of Arabidopsis thaliana to be important for root development and root bacterial communities. At5-MMP is mainly localized in the root vasculature and lateral root, an At5-MMP T-DNA insertion mutant (mmp5 KO) showed reduced root growth and a lower number of root apexes, causing reduced water uptake from the soil. Subsequently, mmp5 KO is sensitive to drought stress. Inhibited auxin transport was accompanied with resistance to indole-3-acetic acid (IAA), 2, 4-dichlorophenoxyacetic acid (2, 4-D), and 1-naphthaleneacetic acid (NAA). The content of endogenous abscisic acid (ABA) was lower in roots of mmp5 KO than in wild type. Genes responsive to ABA as well as genes encoding enzymes of the proline biosynthesis were expressed to a lower extent in mmp5 KO than in wild type. Moreover, drought stress modulated root-associated bacterial communities of mmp5 KO: the number of Actinobacteria increased. Therefore, At5-MMP modulates auxin/ABA signaling rendering the plant sensitive to drought stress and recruiting differential root bacterial communities.
Abbreviations
-
- ABA
-
- abscisic acid
-
- ECM
-
- extracellular matrix
-
- IAA
-
- indole-3-acetic acidx
-
- KO
-
- knock-out
-
- MMP
-
- matrix metalloproteinases
-
- NAA
-
- 1-naphthaleneacetic acid
-
- NPQ
-
- non-photochemical quenching
-
- PCR, polymerase chain reaction; Wt
-
- wild type
-
- WUE
-
- water usage efficiency
1 INTRODUCTION
Proteases play an important role in physiological processes such as cell proliferation, organogenesis, tissue wound healing, and apoptosis (Martinez et al., 2019; Mittal et al., 2016; van der Hoorn & Rivas, 2018). Matrix metalloproteinases (MMPs, EC subclass 3.4.24) belong to the metzincin superfamily and are zinc- and calcium-dependent endo-peptidases (Rawlings & Barrett, 2004) sharing conserved protein structures. In humans, the N-terminal signal peptide of MMPs is responsible for targeting to the secretory pathway, a propeptide domain regulates the enzyme latency and a C-terminal catalytic domain contains the highly conserved active zinc-binding site HEXXHXXGXX(H/D) (Gomis-Rüth, 2003; Klein & Bischoff, 2011). MMPs are synthesized as inactive precursors. Their latency is maintained via chemical coordination between the propeptide domain and the catalytic domain. Their activation, known as the cysteine switch, requires physical delocalization of the propeptide domain from the catalytic site (Hadler-Olsen et al., 2011).
Mammalian MMPs have been shown to function in remodeling of the extracellular matrix (ECM), cell migration, proliferation, and cancer development (Butler & Overall, 2009). Plant MMPs may be involved in growth, development, and immunity (Flinn, 2008; Marino & Funk, 2012). Other plants MMPs were shown to be involved in germination, flowering, programmed cell death, senescence, and stress responses (Das et al., 2018; Li et al., 2015). Nt1-MMP of tobacco (Nicotiana tabacum) and Sl3-MMP of tomato (Solanum lycopersicum) are localized at the plasma membrane (Li et al., 2015; Schiermeyer et al., 2009). Gm1-MMP of soybean is expressed the strongest in mature leaves (Pak et al., 1997), suggesting a role in tissue remodeling, while MMP1 of pine (Pta1-MMP) was expressed at an early stage of embryogenesis, indicating a role in seedling development (Ratnaparkhe et al., 2009). Medicago Mt1-MMP functions as a negative regulator in plant-microbe interaction, over-expressed Mt-MMP1 suppressed nodule formation (Combier et al., 2007). Meanwhile, Os1-MMP in rice was involved in plant development and cellulose/callose depositions (Das et al., 2018). Over-expression of Gm2-MMP in Arabidopsis enhanced high-temperature and humidity stress tolerance, leaves growth, and development as well as seeds vitality and development (Liu et al., 2018).
Arabidopsis thaliana contains five MMPs named At1/2/3/4/5-MMP (Maidment et al., 1999). Recombinant At-MMPs were all shown to be proteolytically active (Marino et al., 2014). While similar preferred cleavage sites were identified for At2/3-MMP and for At1/4-MMP, At5-MMP preferred different amino acids (Marino et al., 2014). So far, no physiological substrates of plant MMPs have been identified and the function of the At-MMPs during plant growth and development is unclear. Gene expression studies showed At2/3-MMP to be down-regulated during various abiotic (ozone, cold, osmotic stress, and salt) and biotic (Botry cinerea, Phytophthora infestans, and Golovinomycis orontil) stresses (Flinn, 2008; Zhao et al., 2017). The At-MMP-encoding genes show high expression during growth and development in seeds/siliques (At1/4-MMP), in young, and developing rosettes (At2-MMP), as well as in all tissues (At5-MMP) (Hruz et al., 2008). An A. thaliana T-DNA insertion line knocking out At2-MMP displayed late flowering and early senescence (Golldack et al., 2002). At2,3,5-mmp triple KO mutants displayed strong resistance to the fungal pathogens B. cinera and G. orontil (Zhao et al., 2017). MMPs therefore participate in several physiological and developmental processes across species, including plant growth, development, and stress responses.
In this study, we demonstrate that At5-MMP knock-out plants (mmp5 KO) display reduced root growth and have less auxin/ABA in their roots compared to wild type. mmp5 KO was resistant to exogenously-added IAA (indole-3-acetic acid), 2, 4-D (2-4-dichlorophenoxyacetic acid), and NAA (1-naphthaleneacetic acid). ABA-responsive and transcription factor genes were down-regulated in mmp5 KO, suggesting mmp5 KO negatively affects drought tolerance. In accordance to its increased leaf stomatal density, mmp5 KO shows increased transpiration rate and stomatal conductance, while stomatal closure was similar to wild type. Root-associated microbiomes differed between wild type and mmp5 KO under drought stress. Therefore, At5-MMP modulates auxin and ABA signaling, promoting drought tolerance, and modulating root microbiomes.
2 MATERIAL AND METHODS
2.1 Plant materials and growth conditions
Arabidopsis thaliana wild type (Col-3) and mutant seeds were sterilized in 75% EtOH for 3 min, in 10% sodium hypochlorite for 5 min and washed with distilled sterilized water three times. They were then kept at 4°C for 2 days. Seeds were plated on Murashige–Skoog media (4.3 g L−1 MS salts [Duchefa], 10 g L−1 sucrose, 8 g L−1 plant agar, pH 5.7). Plates were transferred to a growth chamber (light/dark conditions of 16/8 h at temperatures of 23°C during day and 18°C during night) and grown vertically. MS containing 25 nM IAA (Sigma Aldrich), 100 nM NAA (Sigma Aldrich), 20 nM and 100 nM 2, 4-D (Sigma Aldrich), or 50 μM Kinetin (Sigma Aldrich) were prepared from fresh stocks.
Mutant lines were ordered from the Nottingham Arabidopsis Stock Centre (NASC): At5-mmp (N878060), At1-mmp (N455117), At2-mmp (N439891), At3-mmp (N115923), and At4-mmp (N512824). Homozygous plants deficient in one of the At-MMPs were screened using gene-specific primers. The complementation lines (Comp-1 and -2) were generated after crossing mmp5 KO with plants expressing 35S::At-MMP5 resulting in constitutive expression of At5-MMP. The EMBL/GenBank accession numbers for At1MMP, At2MMP, At3MMP, At4MMP, and At5MMP are NM_117765, NM_105685, NM_102260, NM_130068, and NM_104689, respectively.
2.2 Plant growth conditions for the microbiome experiment
Col-3 was used as wild type (Wt) control, the T-DNA insertion line mmp5 KO (N878060), and the complementation lines expressing At-MMP5 (35S::MMP5-KO, Comp-1, Comp-2) were used to study the microbiome during the watered and drought conditions.
Sterilized seeds were plated on media MS agar media and stratified for 3 days at 4°C and then grown at short day (8 h/16 h photoperiod, 22°C/18°C, and 120 μmol photons m−2 s−1) throughout the duration of the experiment. Fourteen-day-old seedlings were transferred to a soil mixture containing 10% field soil collected from an agricultural field site located in Albany, California, USA (37.8864°N, 122.2982°W), 60% sunshine potting mix, and 30% vermiculite. The plants were potted in 5-cm square plastic plant pots, which were randomized on trays with 15 pots. Five replicates were performed per treatment and genotype. Until the onset of drought treatment, the plants were regularly watered with 750 ml of water per tray. At the age of 4 weeks, the drought treatment was initiated wherein the watered control trays received 750 ml of water and the drought-treated trays received 350 ml of water until the roots (plant age 6–7 weeks) were harvested for DNA extraction and 16S sequencing.
2.3 Sample collection and processing for the microbiome experiment
The plant root samples (five plants per genotype and treatment) were collected by manually extracting the whole plant with the root system from the pot. The excess soil was removed by shaking it off the sterile surface. The soil tightly adhering to the Arabidopsis root surface was severed with ethanol-sterilized forceps and collected in sterile tubes. The samples were quickly frozen in liquid nitrogen and stored at −80°C until further use. Frozen samples were ground to a fine powder using a bead mill while ensuring samples remained frozen. The Qiagen DNeasy Powersoil Kit (Cat No. 12888-100) was used to extract the genomic DNA from all the samples in the experiment.
2.4 Library preparation and amplicon sequence data processing
Library preparation and amplicon sequence data processing was performed according to Simmons et al. (2018, 2020). To account for differences in sequencing read depth across samples, the data per sample were normalized by dividing its reads per ASV by the sum of usable reads, resulting in a table of relative abundance frequencies. To perform alpha-diversity calculations, all sample data were normalized to an even read depth of 12,792 OTUs per sample.
2.5 Microbiome statistical analyses
All 16S statistical analyses were performed in R v3.6.1 (Team, 2013). The Shannon index was determined with the estimate_richness function in the R package phyloseq v1.30.0 (McMurdie & Holmes, 2013), and significance was tested by ANOVA using the aov function in the R stats package. Canonical Analysis of Principal Coordinates (CAP) of Bray-Curtis distance was performed using the ordinate function in the R package phyloseq v1.30.0 (McMurdie & Holmes, 2013). Sample type separation was determined by pairwise PERMANOVA with 1000 permutations using the adonis and calc_pairwise_permanovas functions in the R packages vegan v2.5.6 (Dixon, 2003) and mctoolsr v0.1.1.2 (Carini et al., 2016). Significant differences between Actinobacteria abundances were tested by ANOVA using the aov function in the R stats package, and the Tukey post hoc test using the HSD. test function in the R package agricolae v1.3.1 (de Mendiburu & de Mendiburu, 2020).
2.6 RNA, cDNA synthesis, and transcriptional quantification by RT-PCR
Two weeks old seedlings were used to extract total RNA with the RNase easy kit (Qiagen) according to the manufacturer's instruction. Total RNA was treated with RNase-free DNase (Qiagen) prior to cDNA synthesis. cDNA was synthesized using the iScript cDNA synthesis kit (Biorad) following the manufacturer's instructions. Quantitative RT-PCR was performed using the BioRad CFX 96, with 40 cycles of 10 s at 95°C, 15 s at 60°C and 20 s at 72°C. Reference genes (At-UBIQUTIN and At-TUBULIN) were used to normalize the expression level. Primer sequences are given in Table S1.
2.7 Plasmid construction and plant transformation
At-MMP promoter sequences were fused to pENTR (Invitrogen) and ligated into the gateway binary vector pKGWFS7,0 using LR ligase (Invitrogen). The coding sequences of At-MMPs were fused to pENTR (Invitrogen) and ligated into the gateway binary vectors pGWB5 (Nakagawa et al., 2007) or pK2GW7.0 using LR ligase (Invitrogen). Constructs were transformed into Agrobacterium tumefaciens GV3101. Agrobacterium-mediated flower dipping method was used for plant transformation (Clough & Bent, 1998).
2.8 Auxin and ABA quantification
For auxin measurements, root tips (2 mm from root tip) and upper root parts (2 mm from the hypocotyl-root boundary) from 40 seedlings (5-day-old) were collected in four biological replicates. The auxin measurement was performed as in Andersen et al. (2008) with minor modifications. To analyze the ABA content, roots of plants grown on MS plate for 2 weeks were collected, the ABA content was measured using GC–MS as described by Sundberg (1990).
2.9 GUS assay
Seedlings were incubated in 2 mM X-Gluc (5-bromo-4-chloro-3-indoxy-β-D-glucuronic acid, Fermentas), 0.1% Triton X-100, 50 mM sodium phosphate buffer (pH 7.0), and 1 mM ferri/ferrocyanide and kept overnight at 37°C (Glazebrook & Weigel, 2002).
2.10 Water stress analysis
Wild type and transgenic plants were grown on soil in the growth chamber under short-day conditions (8 h/16 h photoperiod, 22°C/18°C, relative humidity 50%, and 120 μmol photons m−2 s−1) with normal watering for 3–4 weeks before treatment. Long-term water stress analysis was performed by withdrawing watering until the drought effects were observed in the respective genotypes. The experiment was repeated three times to verify the results (Chen et al., 2013).
2.11 Measurements of dehydration
The percentage of dehydration was determined by cutting and weighing well-watered plants as described by Seo et al. (2000) and Koiwai et al. (2004). Ten plants of Col-3 control, the T-DNA insertion line mmp5 KO (N878060), and the line expressing the 35S:: At-MMP5 at the age of 4 weeks were detached and their fresh weight (FW) was measured. Left on the bench at RT, their weight was recorded again after designated time intervals. The relative water content of the leaves was calculated by the formula ([FW-weight at any time point]/FW)*100.
2.12 Stomata size, density, and opening in response to abscisic acid treatment
Stomata aperture measurements were performed on leaves of three to 4 weeks old plants. Using the scotch-tape method, peels from the abaxial side of the leaves were dipped into stomatal opening solution (50 mM KCl, 10 mM MES-KOH, pH 6.2) as described by Lawrence II et al. (2018) for 1 h in a growth chamber with 95 ± 5% RH. Stomata aperture measurements were performed for 1.5 h after the treatment with 10 μmol L−1 abscisic acid (ABA) solution. Control experiments were performed without ABA.
Images of the stomata before and after ABA treatment were taken with a light microscope (Leica DMi8) at ×40 magnification. One leaf (five microscope fields) of five different plants per genotype was evaluated for stomata density and 30 stomatal apertures per leaf were measured using ImageJ software. Standard error and significance at a P-value <0.05 (Student's t test) of the 30 stomatal aperture measurements were calculated.
2.13 Gas-exchange analysis
A portable photosynthesis system (Li-6400xt, Li-Cor) was used to determine the net CO2 assimilation rate (AN) and the stomatal conductance of water vapor (gs) (Tomeo & Rosenthal, 2018). AN was measured at a CO2 concentration (Cr) of 400 μmol mol−1 and a photon flux density of 1000 μmol photons m−2 s−1 on the 7th or 8th leaf of the rosette across five biological replicates per genotype. The leaf chamber temperature was set to 25°C, and the airflow was set to 250 μmol s−1. Gas exchange parameters, including AN, Ci, and gs, were determined during the daytime between 11:00 and 16:00 h. The intrinsic water usage efficiency (WUE) was calculated by the ratio of net photosynthesis (AN) to stomatal conductance of water vapor.
2.14 Chlorophyll fluorescence measurements
After gas exchange measurements, the same leaf was used for chlorophyll a fluorescence (FV/FM) and NPQ measurements. Chlorophyll fluorescence was measured using a pulse amplitude modulation fluorometer (PAM-2500, Walz) as described in the manufacture manual. Fv/Fm was recorded after 30 mins dark adaptation of each leaf disc following the removal of the plant from the growth chamber. A 1-s light saturation pulse of 3000 mol m−2 s−1 was used to record F0. Ten independent biological replicates of 6-week-old whole plants were dark-adapted for 30 mins and the maximum PSII quantum yield Fv/Fm was recorded. The total measuring time was 120 s, with saturation pulses (width = 800 ms); data were collected every 10 s. To determine the NPQ capacity, a light intensity of increasing (PAR) photosynthetically active radiation up to maximum 2000 μmol photons m−2 s−1 was applied (Mishra et al., 2019).
3 RESULTS
3.1 Localization of At5-MMP and its impact on root growth
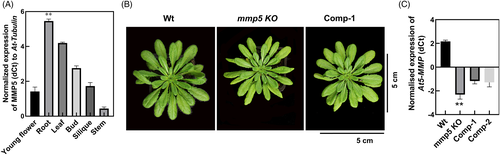
To examine the role of At5-MMP during growth and development, the transcript level of At5-MMP in different tissues like young flowers, roots, leaf, bud, silique, and stem from 3-week-old whole plants were analyzed (Figure 1A). The highest transcript level of At5-MMP was in roots but its expression was ubiquitous in various tissues except for the stem (Figure 1A). The pronounced expression in roots was confirmed using the native At5-MMP promoter fused to the GUS (β-glucuronidase) reporter. At5-MMP promoter activity was observed in the main root and lateral root vasculature (Figure S1A, B and E), but not in the root apex (Figure S1D), epidermal, and cortex cells (Figure S1B and E). At5-MMP promoter expression was also detected in trichomes (Figure S1C). A T-DNA insertion line interrupting At5-MMP was ordered from Nottingham Arabidopsis Stock Centre (NASC) (N878060) and was also complemented with the Wt gene under a 35S promoter (35S::MMP5-KO). T-DNA was inserted into the pro-peptide domain and inactivated the catalytic activity of At5-MMP. Quantitative real-time PCR (qRT-PCR) was performed on RNA extracted from mmp5 KO, Comp-1, Comp-2 (35S::MMP5-KO), and the corresponding wild type plants grown in SD conditions. The relative expression of At5-MMP is reported as dCt values in Figure 1C. The homozygous mmp5 KO displayed significantly lower expression of At5-MMP compared to Wt, while the complementation line showed partial rescue of gene expression. While knocking out At5-MMP resulted in smaller plants (Figure 1B) with significantly reduced rosette weight and leaf length (Figure S2), their rosette sizes and weights (grams) were slightly increased in Comp-1 lines (Figures 1B and S2A). However, there was high variability within the lines; therefore, no significant differences were noted to Wt and the KO line.
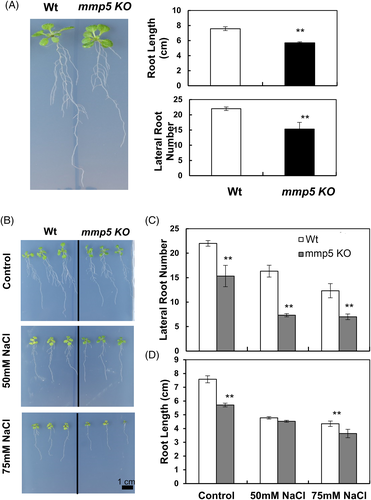
The most obvious change in phenotype between mmp5 KO and the Wt control was reduced root growth (around 30%) and a lower number of lateral roots (Figure 2A). Due to the high homology among the At-MMPs, especially At2/3-MMP and At5-MMP (Figure S3), other MMPs could substitute for the loss of At5-MMP in mmp5 KO. Therefore, root length was also investigated in the other four At-MMP knock-out mutants, but no significant differences in root growth were observed (Figure S4). Thus, At5-MMP seems to be the only At-MMP involved in the regulation of root growth. We determined the effect of the exogenous osmotic stress by treating the seedling with 50 and 75 mM NaCl. mmp5 KO displayed a lower number of lateral roots and reduced root length as compared to the Wt control, indicating substantial sensitivity of the mutant to osmotic stress (Figure 2B,C and D).
3.2 Immp5 KO inhibits auxin transport
The plant hormone auxin (mainly indole-3-acetic acid: IAA) is well known to play important roles in root growth and lateral root development (Overvoorde et al., 2010). The A. thaliana auxin influx carrier mutant aux1 (auxin resistant 1) and auxin efflux carrier mutant pin3 (pin-formed 3) have been shown to have reduced root growth (Friml et al., 2002; Swarup et al., 2008). The aux1 mutant additionally showed an agravitropic root phenotype, reduced lateral root development, and lower IAA accumulation in the root apex caused by inhibited auxin transport (Friml et al., 2002; Marchant et al., 1999; Swarup et al., 2001). Similar to pin3 and aux1, mmp5 KO displayed reduced root growth; however, neither the agravitropic root phenotype nor changes in lateral root development were visible (Figure 2A).
To test whether the reduced root growth of mmp5 KO was caused by inhibited polar auxin transport, auxin measurements were performed within the root (Figure 3A). While no differences in IAA content could be observed in the upper part of the roots, IAA accumulation at the root apex was 23% lower in mmp5 KO compared to Wt. Our hypothesis that auxin content was reduced in mmp5 KO was further substantiated by quantitative real-time PCR (qRT-PCR) to monitor IAA19 gene expression in mmp5 KO and Wt roots. IAA19 is an early auxin-responsive gene (Nakamura et al., 2003). The observed lower expression of IAA19 in mmp5 KO (Figure 3B) was in agreement with the auxin measurement, and indicated that auxin transport from the upper parts of the plant (young leaves and flowers) to the root apex may be inhibited in mmp5 KO. We further examined the mmp5 KO auxin response by exogenously adding indole-3-acetic acid (IAA), or the auxin influx substrate, 2,4-dichlorophenoxyacetic acid (2,4-D). While the root length in Wt was reduced by 16% in the presence of 20 nM 2,4-D compared to normal growth conditions, root length in mmp5 KO was only reduced by 6% (Figure 3C). In the presence of 25 mM IAA, the root length in Wt was reduced by 13%, while the root length in mmp5 KO did not change at all (Figure 3C). The mmp5 KO mutant, therefore, seems to be resistant to 2,4-D and IAA treatments, similar to the resistance shown previously for aux1 (Marchant et al., 1999).
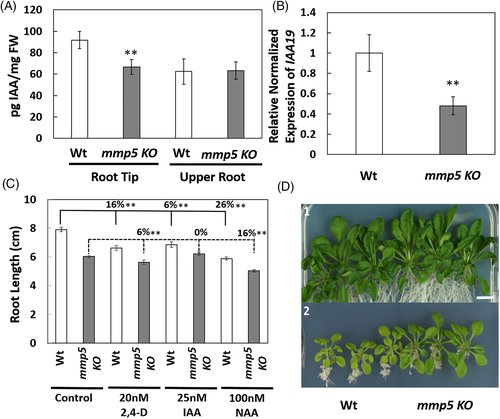
To investigate the auxin transport from root to shoot, mmp5 KO, and Wt were grown in the presence of 2,4-D at high concentration (100 nM) for 3 weeks to monitor their leaf development (Figure 3D). High dosage of 2,4-D application reduces carbon fixation and starch formation and affects mitochondrial respiration and fatty acid β-oxidation in plants (Grossmann, 2010; Romero-Puertas et al., 2004). In the presence of this high dose of 2,4-D, Wt plants displayed reduced leaf development and their leaves were changed to pale green, while leaves of mmp5 KO were green and larger than Wt (Figure 3D). Auxin influx indeed seems to be inhibited from root to shoot in mmp5 KO.
3.3 Reduced expression of At5-MMP increases plant drought sensitivity, transpiration rate, and stomatal density
The root phenotype observed in mmp5 KO plants let us evaluate its drought responses in comparison to Wt and the complementation line (Comp-1). Irrigation was stopped on the soil-grown plants until lethal effects were observed in more than 50% of the replicates of the genotypes under investigation. The mmp5 KO plants showed an early onset of drought sensitivity in comparison to the Wt and Comp-1 line, while the mmp5 KO, Wt, and Comp-1 plants showed consistent healthy growth under controlled growth conditions with regular irrigation (Figure 4A, lower panel). After 12 days of exposure to drought, almost all mmp5 KO plants withered and became chlorotic, whereas most of the Wt and Comp-1 plants continued to grow (Figure 4A, upper panel). After 20 days of drought conditions, all mmp5 KO plants had died, whereas 50–75% of the Wt and the Comp-1 plants survived (not shown).
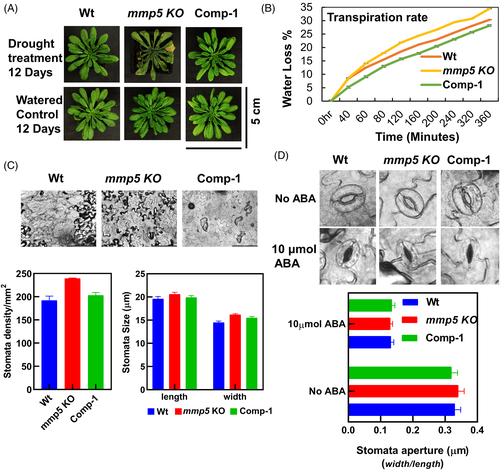
The rate of transpiration was determined by the percentage of water loss. In Wt and mmp5 KO, the rate of water loss was similar at the beginning of the experiment, but a stronger water loss was observed in mmp5 KO compared to Wt and the Comp-1 plants from 1 h onward (Figure 4B). Comp-1, which expresses At5-MMP at a lower level than Wt, displayed a lower transpiration water loss, similar to Wt (Figure 4B).
The average stomatal width-to-length dimension was 19.6 by 14.5 μm for Wt, in contrast to 20.6 by 16.2 μm for mmp5 KO and 19.9 by 15.5 μm for the Comp-1 line (Figure 4C). As there was no significant difference observed between the ratios (width/length) of the stomata across the genotypes, the drought sensitivity displayed by the mmp5 KO plants might be caused by increased stomata density per mm2. The stomatal density of mmp5 KO plants was 240 per mm2, while it was 192 for Wt and 203 per mm2 for the Comp-1 line (Figure 4C lower panel). We further examined the mmp5 KO guard cell apertures in comparison to the ones of Wt and Comp-1 plants at the adaxial side of the leaves under a microscope to understand if the stomatal aperture closure is sensitive to ABA (Figure 4D). Stomatal apertures were determined by calculating the ratio of width/length of the stomata. Stomatal apertures averaged 0.33 μm in Wt, 0.38 μm in mmp5 KO, and 0.37 μm in the Comp-1 line in absence of ABA treatment (Figure 4D). When treated with 10 μmol exogenous ABA, the stomatal apertures of all the genotypes decreased by more than 50%; there was no significant difference between genotypes (Figure 4D). These results suggest the drought sensitivity of mmp5 KO is associated with increased stomatal density rather than sensitivity to ABA.
3.4 Lower amount of endogenous ABA in mmp5 KO causes lower expression of ABA-responsive genes
To overcome drought stress, plants adapt by complex biochemical and physiological processes (Potters et al., 2007). ABA homeostasis is known to be important for resistance against drought stress (Cutler et al., 2010) as well as genetic modifications of signaling compounds (Hauser et al., 2011). Increased ABA content in plants causes stomatal closure and activates down-stream genes to adapt to changed growth conditions (Cutler et al., 2010). Besides this direct effect, ABA was shown to induce the expression of P5CS encoding Δ1-pyrrolline-5-carboxylate synthase, which is involved in proline biosynthesis (Kishor et al., 1995) and even proline has an impact on drought resistance. GC–MS was used to monitor the content of ABA in roots of Wt and mmp5 KO grown at normal growth conditions on MS. Roots of mmp5 KO contained approximately 14% less ABA than Wt (Figure 5A) and additionally, the expression of P5CS was lower in mmp5 KO than in Wt (Figure 5B).
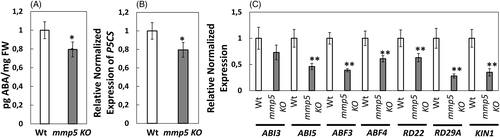
To monitor genes responsive to ABA in mmp5 KO, the expression of transcription factor genes of the ABA-responsive basic leucine zipper (bZIP) were investigated by qPCR (Figure 5C). Transcription of ABI5, ABF3, and ABF4, which are known to bind to the ABA response element (ABRE) (Hwang et al., 2019), were significantly down-regulated in mmp5 KO, while the expression of ABI3 was only slightly reduced (Figure 5C).
3.5 Reduced expression of At5-MMP increases stomatal conductance and lowers WUE without affecting the photosynthetic parameters
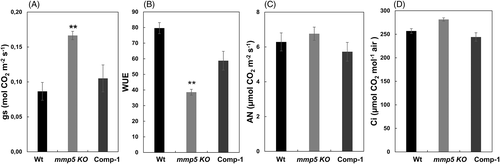
The gas–exchange parameters were first determined under normal growth conditions. No significant differences were observed regarding net photosynthesis AN (μmol CO2 m−2 s−1) and CO2 concentration inside the leaf Ci (μmol CO2 mol−1 air) across the genotypes, whereas leaf stomatal conductance gs (mol CO2 m−2 s−1) was significantly higher in mmp5 KO plants compared to Wt and Comp-1 lines (Figure 6A). The water usage efficiency (WUE intrinsic), that is, the ratio of AN to stomatal conductance of water vapor (AN/gs), showed significantly lower values in mmp5 KO plants in comparison to Wt and the Comp-1 lines (Figure 6B). No significant differences in photosynthetic parameters, that is, net photosynthesis AN (Figure 6C), CO2 concentration inside the leaf (Figure 6D), Fv/Fm or non-photochemical quenching (NPQ) (Figure S5), were observed between the genotypes.
3.6 Reduced expression of At5-MMP leads to recruitment of a stronger drought-responsive bacterial community during drought stress
Mmp5 KO plants display a root growth phenotype and enhanced drought susceptibility. To test if this phenotype has impact on the colonization of root-associated bacteria, Wt, mmp5 KO, and the complementation line Comp-1 (35S::MMP5-KO) were grown in “common garden” trays. Each genotype was grown side-by-side with a shared soil reservoir to ensure equal soil water moisture content between genotypes. Trays were either watered regularly or handled with reduced water treatment, generating modest drought stress. In this way, we ensured the survival of mmp5 KO by visual screening (as in Figure 4A). To evaluate root-associated microbial communities, roots were harvested from 55 days old plants, and 16S rRNA community profiling targeting the V3-V4 variable regions was performed by Illumina MiSeq (Simmons et al., 2018; Xu et al., 2018).
Alpha diversity (within-sample diversity) of root samples did not vary significantly by genotype or treatment (Shannon diversity index, F = 0.177, P = 0.968) (Figure 7A). In contrast, canonical analysis of principal coordinates (CAP) of Bray–Curtis distances constrained for the interaction term treatment by genotype showed a significant beta diversity (between sample diversity; R2 = 0.257, P = 0.01), with CAP1 accounting for 12.2% of the variation, and separating samples by watering treatment (Figure 7B).
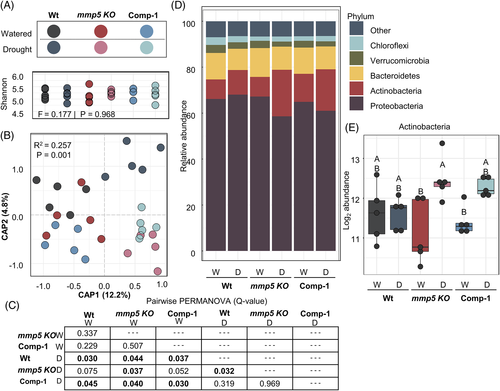
Separation between samples was calculated by pairwise PERMANOVA. No significant difference (P < 0.05) was detected between watered genotypes, suggesting that mmp5 KO does not recruit a unique bacterial community under well-watered conditions. In contrast, each genotype recruited significantly different bacterial profiles between watering and drought treatments (Wt, R2 = 0.205, Q = 0.030; mmp5 KO, R2 = 0.190, Q = 0.037; 35S::MMP5-KO, R2 = 0.204, Q = 0.030), suggesting that plants were responding to water stress (Figure 7C and Table S2). Notably, the drought mmp5 KO community profile significantly differed from Wt (R2 = 0.164, Q = 0.032), indicating that mmp5 KO recruits a unique bacterial community during drought. Additionally, Comp-1 plants partially phenocopied Wt with a community profile that was intermediate between Wt and mmp5 KO (Figure 7B,C).
To understand the underlying cause of beta diversity separation, we evaluated the relative abundance of the top five bacterial phyla. Notably, we observed an increase in the relative abundance of Actinobacteria during drought (Figure 7D), which is a hallmark of drought response in roots (Fitzpatrick et al., 2018; Naylor et al., 2017). To determine the magnitude of this difference observed in Actinobacteria abundance, we determined the sum of Actinobacteria read counts in each treatment. Notably, the largest increase in Actinobacteria occurred between watered and drought mmp5 KO roots (P < 0.05, ANOVA, Tukey-HSD) (Figure 7E), supporting the drought susceptibility phenotypes observed above. Collectively, the 16S microbiome analyses suggest that At5-MMP is not involved in microbial recruitment during well-watered conditions. However, mmp5 KO, which displays a drought susceptibility phenotype, recruits a stronger drought-responsive bacterial community compared with Wt during a modest drought.
4 DISCUSSION
The plant Arabidopsis thaliana contains only five metalloproteinases (At1/2/3/4/5-MMP) compared to mammalian cells that have almost 30. Gene expression studies of the At-MMPs showed that each of the five genes are developmental- and tissue-specific expressed (Flinn, 2008; Maidment et al., 1999). Phylogenetic comparison clustered At2-MMP and At3-MMP, while At5-MMP is more distantly related to these two proteases; At1-MMP and At-4MMP form another cluster with lower identity to At2/3/5-MMP. Even the cleavage site preferences were similar for At2/3-MMP and At1/4-MMP, while the substrate recognition site of At5-MMP seems to be more hydrophobic (Marino et al., 2014). At2/4/5-MMPs contain predicted GPI (glycosylphosphatidylinositol) anchor sites (Flinn, 2008).
In this study, we show the mutant deficient in At5-MMP (mmp5 KO) displays reduced root growth. We demonstrate that At5-MMP is mainly localized in the root vasculature and lateral root. Reduced root growth may be caused by many factors, one of them is the disruption of auxin transport (Petricka et al., 2012). Auxin transport is divided into two distinct pathways: (a) the unregulated bulk flow in mature vasculature and (b) the cell-to-cell carrier-mediated directional transport (Petrášek & Friml, 2009). Cell-to-cell carrier-mediated directional transporters, that is, the plasma-membrane-localized influx (AUX1/LAX1/2/3) and efflux (PINs) carriers, play an important role in controlling auxin transport during development (Friml et al., 2002; Petrášek & Friml, 2009). Mutants impaired in AUX1 or PIN3 expression (aux1 and pin3) have reduced root growth (Friml et al., 2002; Marchant et al., 1999; Swarup et al., 2008). AUX1 is involved in auxin transport from shoot-to-root apex and vice versa and controls auxin unloading in the root apex (Bennett et al., 1996; Marchant et al., 2002; Swarup et al., 2001); in aux1, auxin accumulation at the root apex is diminished compared to Col-0 (Swarup et al., 2001). Additionally, AUX1 can transport 2,4-D along with IAA (Delbarre et al., 1996; Petrášek & Friml, 2009; Yang et al., 2006) and aux1 is resistant to both substrates (Marchant et al., 1999). The auxin transport mutant axr4 (auxin resistance 4) is resistant to the effects of exogenous 2,4-D and displays an agravitropic phenotype similar to aux1 (Dharmasiri et al., 2006). AXR4 is localized at the ER and is required for AUX1, but not PIN1 or 2, to be localized into the plasma-membrane (Dharmasiri et al., 2006). Despite this AXR4 necessity, there is no evidence for an AXR4/AUX1 protein–protein interaction and co-localization (Dharmasiri et al., 2006). Even in mmp5 KO, we observed resistance to 2,4-D, NAA, and IAA. Expression of At-MMP was not observed in epidermal cells (Figure S3), but AUX1 is localized in epidermal cells (Swarup et al., 2001). This may explain why mmp5 KO did not show any agravitropism phenotype (Figure 1B). However, we observed the auxin accumulation at the root apex is lower in mmp5 KO than in Wt, suggesting that reduced root growth in mmp5 KO was caused by an inhibited auxin transport (Figure 2A). To date, no interaction of an auxin influx protein with other proteins has been demonstrated. If AUX1 is a substrate of the At5-MMP protease, its protein level should accumulate in mmp5 KO. The large AUX1 protein contains several cleavage sites shown to be preferred by At5-MMP5 (Marino & Funk, 2012). However, further studies are needed to determine if this interaction occurs. Besides a protein maintenance function, the animal membrane type-1 matrix metalloproteinase (MT1-MMP) propeptide domain acts as an intramolecular chaperone (Cao et al., 2000), the conserved sequence “YGYL” of the propeptide domain plays an important function (Pavlaki et al., 2002). The same conserved propeptide sequence was also found in At5-MMP; it will be interesting to study in Arabidopsis if AUX1 protein activity might be changed after mutagenizing the At5-MMP conserved propeptide domain sequence.
Auxin and ABA are known for root patterning, vascular tissue differentiation, axillary bud formation, and flower organ development (Zhao, 2010). The expression of auxin-responsive genes changes during water stress (Jain & Khurana, 2009). Increased auxin content in plants triggers the growth of lateral roots (root biomass) and these lateral roots contain more root apices (Shi et al., 2014). Plants with an increased number of lateral roots are known to tolerate enhanced drought stress, while the lower number of lateral roots observed in mmp5 KO induces severe drought stress (Figure 1B). An Arabidopsis mutant over-expressing the gene YUCCA7 encoding a flavin monooxygenase displays an increase amount of endogenous auxin and increased resistance to drought stress (Lee & Luan, 2012). In contrast, over-expressing GH3-2 encoding an auxin-amido synthetase in rice reduced the amount of endogenous auxin, resulting in decreased drought stress tolerance (Du et al., 2012).
ABA and auxin are both important factors, which influence each other, in early developmental stages (seed germination and seedling development) (Liu et al., 2007). The expression of ABA-responsive genes (RD22, RD29A, RD29B, and DREB2A) was found to be higher in plants with increased endogenous auxin (Yamaguchi-Shinozaki & Shinozaki, 2006). Plants treated with an inhibitor of ABA biosynthesis, Fluridone, contain less endogenous auxin, and their auxin biosynthesis genes are down-regulated (Du et al., 2013). Roots of mmp5 KO contained less endogenous auxin, consistent with the observation that the expression of the ABA-responsive genes was lower in the mutant than in Wt (Figure 5). The expression of ABI5 was lower in mmp5 KO, but it was not the case for ABI3. ABI3 might coordinate ABA and auxin signaling. The B3-binding domain of ABI3 has conserved sequences, similar to the auxin response factor (ARF) genes. ARF genes are thought to interact with the plant hormone indole-3-acetic acid (IAA) to modulate the auxin response; however, an abi3 mutant did not show stimulated lateral root initiation after auxin treatment (Brady et al., 2003; Tiwari et al., 2001). The ABA content in abi3 is not changed, while abi5 (and mmp5 KO [Figure 5A and C]) contain less ABA than wild type (Verslues & Bray, 2006). While we can speculate that At5-MMP directly influences ABI5 independently from the ABA signaling pathway, an indirect pathway, via the auxin-level, is more likely. Lower ABA content and less number of lateral roots decrease the resistance against drought stress. Franks and Farquhar (2001) showed ABA-sprayed leaf had higher stomata density compared to untreated leaf without significant effect on photosynthesis activity, which means there is no connection between stomata number and photosynthetic capacity. However, stomata density is highly correlated with the transpiration rate, as we demonstrate for mmp5 KO (Figure 4B, C and D). Although the molecular mechanism affecting the stomatal density in mmp5 KO remains unclear, our findings resonate with those reported by Berger and Altmann (2000) and Von Groll et al. (2002) showing that another extracellular protease, SUBTILISIN-LIKE SERINE PROTEASE SDD1, is involved in the regulation of stomatal density and distribution. Taken together, stomata density, number of lateral roots, and root length are essential to drought stress resistance.
It is still difficult to explain the explicit mechanism by which At5-MMP influences root length, ABA biosynthesis, stomata density, and drought sensitivity. We wanted to determine if these phenotypes would collectively affect the accumulation of root-associated bacterial communities during water deficit in the model plant Ai carrying a T-DNA insertion in the At5-MMP gene. Previous works have shown the enrichment of Actinobacteria within the root microbiome of angiosperms (Fitzpatrick et al., 2018; Naylor et al., 2017) during drought stress. Therefore, we hypothesized that mmp5 KO plants would have increased Actinobacteria abundance during drought stress. The phytohormone ABA, which plays an important role during drought stress tolerance in plants, is present in the rhizosphere, where it is metabolized by bacteria and may help the plants to cope with the stress or modify the composition of bacterial communities (reviewed by (de Vries et al., 2020, Xu & Coleman-Derr, 2019). The lower levels of ABA, stunted root growth, and reduced drought-tolerant traits observed in mmp5 KO mutants do not have an obvious effect on the bacterial alpha diversity. The samples did not vary significantly by genotype or treatment (Figure 7A) but the canonical analysis of principal coordinates (CAP) of Bray–Curtis distances showed a significant beta diversity with CAP1 accounting for 12.2% of the variation. Samples were separated by watering treatment (Figure 7B), indicating that mmp5 KO mutants during drought stress recruit a unique bacterial community. While dissecting the underlying cause of beta diversity separation, we observed increases in Actinobacteria in mmp5 KO drought samples compared to the watered mmp5 KO samples. An enrichment of Actinobacteria in mmp5 KO drought samples is in agreement with the drought-susceptible phenotype showed by the mutants that occur taxonomically across various plant hosts (Edwards et al., 2018; Fitzpatrick et al., 2018; Naylor et al., 2017; Santos-Medellín et al., 2017). To conclude our study shows that mmp5 KO does not recruit unique microbial communities during well-watered conditions, but accumulates an altered root-associated bacterial community during water deficit.
Reduced ABA promotes increased drought susceptibility due to a lower water-use efficiency and phenotypical changes susceptible to drought (Daszkowska-Golec, 2016). In agreement with the aboveground phenotypes, a modest drought-induced shifts in bacterial community structure in mmp5 KO plants, whereas no significant shift was observed in Wt or complementation plants. This suggests the mmp5 KO microbiome is responding to the perception of drought in the roots rather than soil moisture content, as the experiment was performed in a “common garden.” In other words, mmp5 KO plants are more susceptible to drought, and this susceptibility leads to changes in root-associated microbiome consistent with a plant responding to drought. Further studies are needed to investigate how At5-MMP is involved in root development together with auxin and ABA regulation and how At5-MMP inactivation regulates the recruitment of unique microbial communities during drought stress. The identification of MMP5 substrates will provide vital information on how At5-MMP regulates developmental processes in plants.
ACKNOWLEDGMENTS
We acknowledge financial support by the Swedish Research Council VR (grant number 2019-04472 to CF), the Kempe Foundation for SYK's Post-doctoral fellowship and Umeå Universitet supporting LSM's UC Berkeley Visiting Scholar programme under the Berkeley Global engagement scheme. The Swedish Metabolomics Centre at Umeå is acknowledged for technical support analyzing ABA and auxin and Dr. Giada Marino for providing At5-MMP promoter-GUS seeds.
AUTHOR CONTRIBUTIONS
Sung-Yong Kim, Laxmi S. Mishra, and Daniel F. Caddell performed the experiments, were involved in the formal analysis and characterized the mutant plants. Devin Coleman-Derr and Christiane Funk supervised the project. Christiane Funk conceptualized the project and was responsible for funding acquisition. All authors were involved in writing the manuscript.
Open Research
DATA AVAILABILITY STATEMENT
Data submission for Microbiome experiment: Prior to publication, all datasets and scripts for 16S analyses are available through github (https://github.com/colemanderr-lab/Mishra-2020) and all short read data has been submitted to the NCBI and can be accessed through BioProject PRJNA655744.




