The res (restored cell structure by salinity) tomato mutant reveals the role of the DEAD-box RNA helicase SlDEAD39 in plant development and salt response
C. Capel and I. Albaladejo should be considered joint first authors.
Funding information: Secretaría de Estado de Investigación, Desarrollo e Innovación, Grant/Award Numbers: AGL2015-64991-C3-1-R, AGL2015-64991-C3-2-R, AGL2015-64991-C3-3-R, AGL2017-88702-C2-1-R
Abstract
Increasing evidences highlight the importance of DEAD-box RNA helicases in plant development and stress responses. In a previous study, we characterized the tomato res mutant (restored cell structure by salinity), showing chlorosis and development alterations that reverted under salt-stress conditions. Map-based cloning demonstrates that RES gene encodes SlDEAD39, a chloroplast-targeted DEAD-box RNA helicase. Constitutive expression of SlDEAD39 complements the res mutation, while the silencing lines had a similar phenotype than res mutant, which is also reverted under salinity. Functional analysis of res mutant proved SlDEAD39 is involved in the in vivo processing of the chloroplast, 23S rRNA, at the hidden break-B site, a feature also supported by in vitro binding experiments of the protein. In addition, our results show that other genes coding for chloroplast-targeted DEAD-box proteins are induced by salt-stress, which might explain the rescue of the res mutant phenotype. Interestingly, salinity restored the phenotype of res adult plants by increasing their sugar content and fruit yield. Together, these results propose an unprecedented role of a DEAD-box RNA helicase in regulating plant development and stress response through the proper ribosome and chloroplast functioning, which, in turn, represents a potential target to improve salt tolerance in tomato crops.
1 INTRODUCTION
Modern agriculture is facing its greatest challenge due to climate change, which is already affecting global agricultural systems. Therefore, the development of tolerant crops is essential for a sustainable agriculture in order to mitigate yield loss caused by climate change. To achieve an improved abiotic stress tolerance, it is a priority to identify key genes involved in such tolerance. A gene family that is gaining attention in abiotic stress tolerance is the one constituted by RNA helicases (Baruah, Debbarma, Boruah, & Keshavaiah, 2017; Nawaz & Kang, 2017; Nidumukkala, Tayi, Chittela, Vudem, & Khareedu, 2019; Owttrim, 2013). Most RNA helicases (RHs) are classified into two super-families, SF1 and SF2, according to their sequence and structural features. The DEAD-box family is the largest group of SF2 helicases, and they are characterized by the presence of nine conserved amino acid motifs. Motif II of SF2 proteins is characterized by the presence of the amino acid sequence, Asp-Glu-Ala-Asp (DEAD), hence these proteins are named DEAD-box helicases. Apart from these motifs, DEAD-box proteins show different N- and C-terminal extension sequences, which presumably determine their substrate specificity, subcellular localization or their interaction with other partners (Byrd & Raney, 2012; Fairman-Williams, Guenther, & Jankowsky, 2010).
Increasing numbers of reports have pointed to the importance of DEAD-box RHs as regulators of RNA metabolism in chloroplasts and mitochondria. Thus, a variety of nucleus-encoded RHs are targeted to chloroplasts or mitochondria and play key roles in RNA processing, intron splicing and translation, and, therefore, they are essentially required for organellar biogenesis and functionality, as well as stress responses (Nawaz & Kang, 2017). Thus, DEAD-box RHs targeted into chloroplasts or mitochondria, such as RH3 and ABO6, regulate ABA or auxin signaling-mediated stress pathways (Asakura, Galarneau, Watkins, Barkan, & van Wijk, 2012; Gu, Xu, Lee, Lee, & Kang, 2014; He et al., 2012). The chloroplast-localized RH OsABP is involved in the response of rice to different abiotic stresses (Macovei, Vaid, Tula, & Tuteja, 2012), and mitochondria-localized OsSUV3 improves the photosynthesis, together with the antioxidant systems, in rice, under salinity (Tuteja, Sahoo, Garg, & Tuteja, 2013). More recently, it has been proved that BrRH22, a chloroplast-targeted DEAD-box RH from Brassica rapa, positively contributes to the response to abiotic stress of transgenic Arabidopsis by shaping translation of chloroplast genes through its RNA chaperone activity (Nawaz, Lee, Park, Kim, & Kang, 2018). Although these previous studies have clearly pointed to the importance of chloroplast-targeted DEAD-box RHs during stress responses, our current knowledge on the stress-responsive roles of chloroplast-targeted DEAD-box RHs remains limited.
Tomato (Solanum lycopersicum) is one of the most widely produced crops and it is also considered a model species for the study of plant development and abiotic stress tolerance (Zouine et al., 2017). Forty-two genes encoding DEAD-box RHs have been identified in the genome of tomato (Cai et al., 2018; Xu, Zhang, Lu, Cao, & Zheng, 2013), and although the function of most of them remains unknown, these may play important roles in the genetic control of plant development and in the response to biotic and abiotic stresses as proposed in other plant species (Nidumukkala et al., 2019). Recently, Zhu et al. (2015) reported the identification and characterization of two tomato nuclear-localized DEAD-box RHs, SlDEAD30 and SlDEAD31, and their results suggest that SlDEAD31 is a positive regulator of salt and drought tolerance. As part of a project on tomato functional genomics, we previously identified a recessive mutant, named res (restored cell structure by salinity), which showed a chlorotic phenotype from the cotyledon stage, remarkable plant growth inhibition, and important alterations in cell and chloroplast structures. One of the most interesting features of the res mutant was that salt stress normalizes its plant development, since, under salt-stress, the phenotypic alterations of the res mutant disappear and the chloroplast structure is recovered (García-Abellán et al., 2015). Through a whole-genome expression analysis, we found that the phenotype normalization of the res mutant growing under salt stress conditions was associated with an important transcriptome reprogramming (Albaladejo et al., 2018). Among those transcriptome changes, the ARS1 transcription factor, which is involved in salt tolerance by regulating stomatal closure during stress (Campos et al., 2016), was specifically salt-upregulated in res leaves as compared to wild-type ones. In addition, the specific salt-upregulation of key genes involved in photosynthesis might explain the enhancement of photosynthetic efficiency and phenotype recovery in res mutant (Albaladejo et al., 2018). In this research work, the molecular cloning and functional characterization of the RES gene are reported. RES codes for the DEAD-box RNA helicase, SlDEAD39, and the functional role of this chloroplast-targeted DEAD-box RH in chloroplastidial 23S rRNA processing is proven. Finally, we provide evidence supporting that the function of SlDEAD39 is crucial for chloroplast structure and function under non-stressful culture conditions but not under salt-stress condition, where the chloroplastidial 23S rRNA processing is partially recovered, and therefore are chloroplast function and structure. A possible molecular mechanism to explain the recovery of 23S rRNA processing under salinity is discussed.
2 MATERIAL AND METHODS
2.1 Plant material
The res mutant was isolated from a collection of insertional mutants generated in the Moneymaker cultivar of tomato (Solanum lycopersicum L.) (Pérez-Martín et al., 2017). Phenotypic characterization, recessive inheritance and somaclonal origin of the recessive res mutant were previously described (García-Abellán et al., 2015). A T3 line, segregating for the res mutant phenotype and lacking any T-DNA sequence (García-Abellán et al., 2015), was used in the co-segregation analysis. From the T4 progeny, a homozygous mutant line was selected for further gene expression analysis. A single plant of this homozygous res line was crossed with a single plant of S. pimpinellifolium accession, LA1589, and a descendant F1 plant was self-pollinated, and the F2 segregating generation was used for mapping and cloning the RES gene.
2.2 Plant growth and salt stress assays
Seeds were germinated in darkness, in a 2:1 (v/v) mixture of peat:perlite, at 28°C temperature and 90% of relative humidity. After emergence, plants were grown in a controlled growth chamber with 16 hr light/8 hr darkness photoperiod, with light of a photosynthetic photon flux (400–700 nm) of 350 μmol m−2 s−1 at the plant level, provided by fluorescent tubes (Philips Master TL-D 58 W/840 REFLEX, Holland), 25°C of temperature and 50–60% relative humidity. During this period, plants were irrigated daily with half-strength Hoagland solution (Hoagland & Arnon, 1950). Plants were grown in these controlled culture conditions until the desired developmental stage for each experiment was achieved.
Analysis of the spatial expression pattern of SlDEAD39 was carried out in wild type (WT) and res mutant adult plants (20–21 fully developed leaves) grown in 10 L plastic pots filled with peat. Samples of roots, young and mature leaves (first and fourth fully developed, respectively), stems, flowers and immature fruits were taken from individual plants. For each sample, three biological replicates were taken by pooling the material from three individual plants that were immediately frozen with liquid nitrogen and conserved at −80°C.
For short-term hydroponic salt treatment experiments, seedlings with two fully developed leaves were transferred to a hydroponic culture system. Plants were grown hydroponically in an aerated half-strength Hoagland solution as previously described (García-Abellán et al., 2015). Salt stress was applied to nine plants per genotype (WT, res and transgenic plants) at the stage of eight fully developed leaves and consisted of half-strength Hoagland solution supplemented with NaCl (Panreac). An equivalent number of plants were fertirrigated with half-strength Hoagland solution in the absence of salt as control. In order to avoid osmotic shock, salt treatment was applied in two steps, first 100 mM NaCl for 12 hr and then the NaCl concentration was increased until 200 mM and maintained up to the end of the experiment.
Long-term salt treatment was carried out in a greenhouse located at the campus of the University of Murcia (Espinardo, Region of Murcia, Spain). The temperature was programmed to daily oscillate between 15°C (night) and 28°C (day), and relative humidity was maintained at 60%. At the fourth-leaf stage (30 days after sowing), 18 plants per genotype (WT and res mutant plants) were transplanted to plastic pots containing 17 L of coco peat, using a drip irrigation system, with 3 L h−1 drippers. The fertigation solution (Hoagland solution, Hoagland & Arnon, 1950) was prepared in 2,000 L tanks with local irrigation water (EC = 0.9 dS m−1), and pH and EC were regularly monitored. At the 10th-leaf stage, salt treatment was applied to half of the plants of each genotype for 80 days, by means of a fertigation solution supplemented with 100 mM NaCl, while the other half was irrigated without salt (control condition). Salt concentration was selected on the basis of previous studies (Campos et al., 2016; Egea et al., 2018). Ripe fruits of each plant were collected from approximately the 40–80th day period of salt treatment and total weight of fruits per plant (yield), number of fruits per plant and the average fruit weight were recorded to estimate fruit production.
2.3 DNA extraction and molecular marker analyses
Genomic DNA was purified and quantified by comparison with DNA standards after electrophoresis in 1% agarose gels in 1× SD buffer as previously described (Capel et al., 2015). Genetic markers were selected from a published tomato genetic map and were genotyped as described (Capel et al., 2017). Genomic sequences were obtained from a DNA pool constructed by mixing equal amounts of DNA from 16 F2 homozygous res mutant plants. The DNA was used for library generation using the Illumina TruSeq DNA protocol and sequencing was performed in an Illumina HiSeq2000 platform (Illumina, Inc., USA) with 150 pb paired-ends. Single-nucleotide polymorphisms and insertions/deletions were identified by comparison with the tomato reference genome using GenomeMapper (Schneeberger et al., 2009). The mutated nucleotide in SlDEAD39, identified in res mutant plants, generates a restriction site for PfeI restriction enzyme. Thus, the co-segregation analysis performed consisted the amplification of the polymorphic region by PCR, using primers 339genoF and 339genoR (Table S1), in plants, from a T4 segregating population, for the res mutation, followed by the enzymatic digestion of the PCR product, with PfeI, and the electrophoretic separation of the restriction fragments.
2.4 Quantitative real-time PCR analysis
All primers used for RT-qPCR are listed in Table S2. Frozen tissues were grounded in a mortar and approximately 200 mg of powder was used for total RNA extraction with Trizol reagent (Invitrogen). Contaminant DNA was removed with RNase-free DNase (DNA-free kit, Ambion, Austin, TX) and RNA quality was analyzed by electrophoresis on a denaturing agarose gel. Total RNA was quantified in a Nanodrop2000c spectrophotometer (ThermoFisher Scientific). Approximately 3 μg of RNA was used for cDNA synthesis using the First Strand cDNA Synthesis Kit (ThermoFisher Scientific). Finally, quantitative PCR was performed in a 7,300 Real-Time PCR System (Applied Biosystems, Foster City, CA) using the SYBR Green PCR Master Mix (Applied Biosystems) following the manufacturer guidelines. In each reaction, 1 μl of undiluted cDNA and 3 μM of forward and reverse primers were used. All reactions were performed in three biological replicates. Gene expression was quantified using the comparative method (2–ΔΔCt) (Schmittgen & Livak, 2008). Relative expression data were calculated from the difference in threshold cycle (ΔCt) between the studied gene and the housekeeping gene, SlEF1α (acc. AB061263). Regarding the analysis of SlDEAD39 expression during salt stress, calibration was made using the expression level of WT in the absence of salt stress in each tissue.
2.5 Generation of transgenic tomato lines
The complete open reading frame of SlDEAD39 was obtained by RT-PCR performed with primers listed in Table S1. PCR fragments were cloned in the binary vector, pRokII (Baulcombe, Saunders, Bevan, Mayo, & Harrison, 1986), to generate a 35S::SlDEAD39 overexpression gene construct. This gene construct was used for both analyzing phenotype effects of SlDEAD39 constitutive expression and molecular complementation of res mutant. An RNA interference (RNAi) approach was used to develop SlDEAD39 silencing lines. With this aim, a genomic PCR fragment of the SlDEAD39 gene was amplified with primers indicated in Table S1, and was cloned in sense and antisense orientations separated by intronic sequences into the pKannibal vector (Wesley et al., 2001) to generate a pKannibal-SlDEAD39 plasmid. The resulting plasmid was digested with NotI and the entire product was cloned into the binary vector pART27 (Gleave, 1992). Agrobacterium-mediated transformation was carried out from WT and res mutant explants and only diploid transgenic lines were selected as previously described (Campos et al., 2016). Transgenic lines were firstly characterized under in vitro conditions using a half-strength Murashige-Skoog medium (Murashige & Skoog, 1962) supplemented with mineral salts (Duchefa), sucrose (10 g l−1) (Panreac) and indole-3-acetic acid (1 mg l−1) (Sigma-Aldrich). Subsequently, these lines were acclimated to obtain seeds and to characterize in vivo development and salinity response.
2.6 Northern blot analysis
RNA-blot analyses to study 23S rRNA processing were carried out from WT and res plants grown in normal conditions (control) and after a period of salt treatment when phenotypic normalization was observed (5 days 200 mM NaCl). Total RNA was extracted from the youngest developed leaf and its stem, as described above for quantitative real-time PCR analyses. Then, RNA was electrophoresed and transferred onto Hybond-N+ nylon membranes (GE Healthcare), following the protocol described in Ausubel et al. (1993). Primer pairs used, to generate probes for different regions of the rRNA, are listed in Table S3; PCR products were cloned and sequenced to confirm the identity of the cloned fragment. Radiolabeled probes were generated using α-32P-dCTP (Perkin Elmer), the High Prime DNA Labelling Kit (Roche) together with the insert of each selected clone as template.
2.7 RNA binding analysis of SlDEAD39 protein
The complete open reading frame of the SlDEAD39 gene was amplified from the binary vector, pRokII, with primers listed in Table S1 and cloned in the pGEX4T-3 expression vector. The resulting plasmid was introduced into E. coli BL21-CodonPlus cells (Agilent) which, in turn, were grown to the exponential phase when the protein expression was induced by addition of IPTG to 1 mM and further incubation for 16 hr at 20°C. After bacteria disruption by sonication, protein purification was performed using reduced GST-sepharose (Pharmacia) according to the manufacturer's instructions. The fusion protein was digested with thrombin, and the SlDEAD39 protein was eluted using 20 mM Tris–HCl buffer (pH 8.6), 100 mM NaCl, 0.3 mM CaCl2, 1 mM DTT and conserved at −20°C after addition of 50% glycerol. The RNA binding analysis was performed as described in Nishimura, Ashida, Ogawa and Yokota (2010) with synthetic ribonucleotides from the tomato 23S transcript, RNA5 5′- GCCCCGUAACUUCGGGAGAAGGGGUGCCUCCUCACAAAGGGGGU -3′ and RNA2 5′-UGCCUCCUCACAAAGGGGGUC-3′. The mobility of the complex was determined as described in Rio (2014).
2.8 Microscopy analyses
In six independent plants of WT, res mutant, RNAi-9 silencing line and C-2 complementation line, leaflet sections of 1.0 mm2 from the first fully expanded leaf were collected. For white light microscopy analysis, fixation, dehydration, resin infiltration, preparation of sections and their staining from the polymerized blocks were performed as described in Albaladejo et al. (2017), and the stained sections were observed under a Leica LMR light microscope (Leica Microsystems, Wetzlar, Germany). Transmission electron microscopy analysis was performed as we previously described (García-Abellán et al., 2015) using a Philips Tecnai 12 transmission electron microscope (Philips, Eindhoven, The Netherlands) equipped with a CCD SIS MegaView III camera. Reagents used in the microscopy analyses came from Electron Microscopy Sciences (Hatfield, PA).
2.9 Measurements of physiological parameters
Chlorophyll content and fluorescence emission of chlorophylls were analyzed in the first fully developed leaf of WT, res mutant and transgenic overexpression, complementation and silencing lines by means of a portable device, SPAD-502, that measures chlorophyll fluorescence (Minolta, Kyoto, Japan) and a portable chlorophyll fluorometer OS-30p (Opti-Sciences, Hudson USA), respectively, according to García-Abellán et al. (2015).
The content of different sugars was analyzed in the fully developed leaves closer to the fruit trusses of WT and res mutant plants. Aliquots of frozen samples were lyophilized over 48 hr and then kept at −20°C in a closed recipient with silica gel. The extraction protocol and 1H NMR analysis were performed according to Massaretto et al. (2018). Measurements were carried out at the Metabolomics Service of CEBAS-CSIC (Murcia, Spain).
2.10 Statistical analysis
Experimental data are presented as mean ± standard error (SE) values of three biological replicates of six plants each per genotype and treatment. Statistical analysis was performed by Student's t-test and LSD, which were applied to identify significant differences among mean values at p < .05, using the SPSS 24.0 software package. Significant differences between means are denoted by asterisks or letters.
3 RESULTS
3.1 The res phenotype is due to the mutation in the coding sequence of SlDEAD39 gene
Tomato plants homozygous for the res mutation showed an evident chlorosis and severe damages affecting cell structure and organization, which, together, caused growth inhibition during vegetative development (García-Abellán et al., 2015). The conspicuous alterations of the res mutant provide a robust phenotype for fine mapping of the RES locus. With this end, a genetic analysis was performed from a F2 segregating population generated from the cross between a res mutant plant and a plant from S. pimpinellifolium accession, LA1589. A single F1 plant was self-pollinated, and in the F2 segregating population, 252 plants showed a WT phenotype, whereas 71 plants showed the characteristic res mutant phenotype. Chi-square test (χ2 = 1.57, p = .21) supports the recessive genetic inheritance of the res mutation in this population. F2 plants were genotyped using molecular markers distributed along the 12 chromosomes selected from a tomato genetic map (Capel et al., 2017), which allowed us to locate the RES locus on the long arm of chromosome 12. Fine mapping delimited the RES gene into an interval of 654 Kb between the SOC (Solyc12g056460) MADS-box gene and the ETR2b gene (Solyc12g056990), as shown in Figure 1a. This genomic region contains 52 predicted genes, most of them with putative biological functions that could be related to the res mutant phenotype.
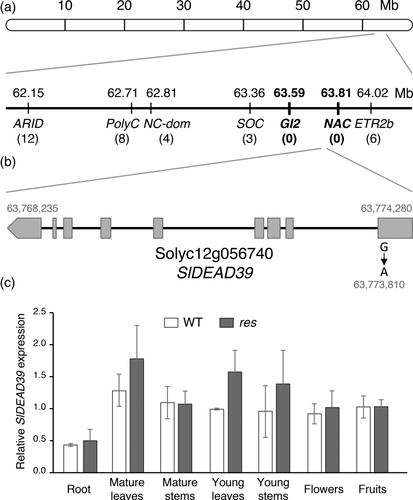
In parallel, we obtained a 30-fold coverage of the tomato genome by sequencing a pool of DNA from 16 F2 homozygous res mutant plants. Within the 654 Kb mapping interval, a single nucleotide difference was identified in the res pooled mutant DNA with respect to the tomato reference genome sequence (The Tomato Genome Consortium, 2012), concretely a 63,773,810 G > A point substitution located in the first exon of the Solyc12g056740 gene (Figure 1b). The presence of this mutation was confirmed by dideoxy sequencing in res mutant plants, and then a specific genetic marker for this mutation was designed and mapped in the whole F2 population. Results showed that G > A mutation co-segregated with the res mutant phenotype of individual plants forming the mutant pool (Figure S1).
An additional genetic analysis of a res segregating population, consisting of 114 individuals, was carried out in order to confirm that the mutation localized in the Solyc12g056740 gene was responsible for res phenotype. Although a lower than expected (1/4) number of individuals with res phenotype was found in this population (104 WT:10 res), this analysis confirmed the recessive inheritance of the res mutation and indicated that the mutation could cause a loss of seed viability over time. Despite this, genotyping of all these plants confirmed that all res mutant plants were homozygous for the mutant allele, whereas WT individuals were either homozygous or heterozygous for the WT allele, hence the mutant phenotype co-segregated with the G > A mutation at the Solyc12g056740 locus. These results supported the causality of the res mutant phenotype and led to preliminarily naming the gene affected by the mutation described above as RES.
Expression analyses showed that RES transcripts were accumulated in all vegetative tissues (leaves, stems and even roots) and reproductive organs (flowers and fruits) of WT plants, and at quite similar levels than in res mutant plants (Figure 1c). According to the tomato genome database Sol Genomics (Bombarely et al., 2011), the Solyc12g056740 (RES gene) encodes the SlDEAD39 protein, one of the 42 DEAD-box RNA helicases (RH) of tomato. SlDEAD39 contains 636 amino acids, its predicted molecular mass is 70 kDa and its isoelectric point is 9.88. However, the biological function of the tomato, SlDEAD39, protein has never been reported. The most homologous Arabidopsis protein to SlDEAD39 is AtRH39 and a sequence alignment performed using Clustal Omega tool revealed a 65% of sequence identity and 75% similarity (Figure S2). The nine characteristic motifs of DEAD-box proteins are highly conserved between AtRH39 and SlDEAD39. Moreover, both proteins contain a predicted N-terminal plastid transit peptide (Figure S2), suggesting that SlDEAD39 should be located in the chloroplast as it was previously found for AtRH39 (Nishimura et al., 2010). Mutation at res caused a conserved Gly residue located in motif I to be replaced by Asp (Figure S2) and, therefore, it might not be a null allele but a loss-of-function allele responsible for the res mutant phenotype. This Gly residue is conserved among very diverse plant species, suggesting it plays an essential role in the functionality of DEAD-box RHs (Nishimura et al., 2010).
3.2 SlDEAD39 silencing plants phenocopy the res mutant
In order to demonstrate that the identified mutation is the sole responsible for the res mutant phenotype, nine independent diploid transgenic lines silencing SlDEAD39 were generated using an RNA interference strategy (RNAi). The phenotypic analyses showed that differences between these RNAi lines could be associated with the different levels of SlDEAD39 gene silencing observed in these lines when they were grown in vitro (Figure S3). Thus, seven RNAi lines with relative SlDEAD39 expression levels of 0.1–0.9 (i.e. 2, 7, 8, 9, 10, 11 and 14) showed similar phenotypes similar to the res mutant phenotype, although one of them (RNAi-2) showed a slight slower plant development, and necrotic zones appeared in its leaves (Figure S4). However, two of RNAi lines (21 and 34), with undetected levels of SlDEAD39 transcripts, showed extreme chlorotic phenotypes when they were grown in vitro culture, with impaired vegetative development, and they finally died (Figure S4).
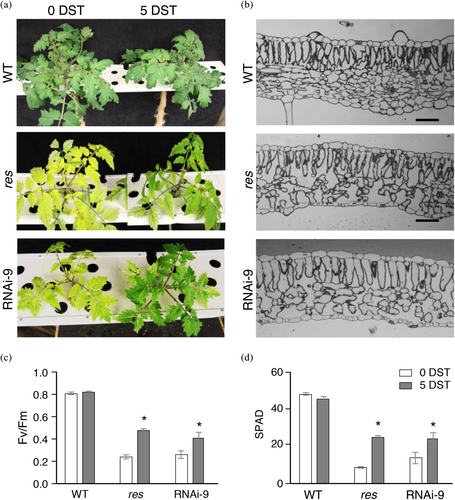
After the RNAi lines were acclimated and grew under in vivo control conditions, the RNAi-9 plant showed a phenotype similar to res mutant plants in terms of plant growth, leaf chlorosis and cell structure alterations (Figure 2a,d). However, RNAi-2 showed a much more severe phenotype, with markedly reduced development and greater chlorosis compared to RNAi-9 and res mutant plants (Figure 2a,b). Moreover, RNAi-2 presented symptoms of leaf senescence (Figure 2c) more evident in the leaflets borders where chlorophyll content was lower. Finally, such abnormalities caused the death of the plant after a few weeks of acclimation in vivo, indicating that SlDEAD39 gene is essential for the proper development of tomato plants. These results also indicate that res is not a null allele since mutant plants are able to continue their development, although with severe alterations.

One of the most intriguing features of res mutant was the disappearance of developmental abnormalities mentioned above when plants were grown under salt stress; indeed, leaf chlorosis and cell structure alterations were completely restored in salinity conditions (García-Abellán et al., 2015). In this research, we wondered whether RNAi lines also normalized their phenotypes upon salinity, and for this propose the RNAi-9 line showing a phenotype similar to res mutant plants was selected for further characterization under control and salt conditions. Interestingly, the salt responses of res mutant and RNAi-9 line were similar after 5 days of salt treatment (DST), as both genotypes began to show greener leaves and normalization of the cell structure alterations (Figure 3(a),(b)). Next, we measured chlorophyll fluorescence (Fv/Fm) and chlorophyll content (SPAD) in WT, res and RNAi-9 leaves, and we observed that values of both parameters significantly increased in leaves of res and RNAi-9 plants after 5 DST, coinciding with the beginning of the phenotypic normalization (Figure 3c,d).
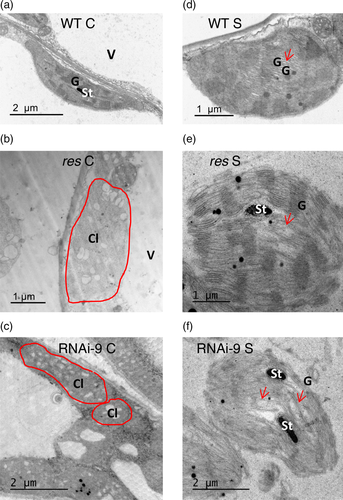
Transmission electron microscopy (TEM) analyses demonstrated that chloroplasts of res mutant showed an altered ultrastructure (García-Abellán et al., 2015). Given the phenotype similarity of the RNAi-9 line with the res mutant, we wonder if its chloroplasts ultrastructure was similar. In order to check this issue, the ultrastructure of chloroplasts was analyzed by TEM not only in WT and RNAi-9 but in res mutant too. The results clearly showed that the ultrastructure of the RNAi-9 chloroplasts, like those of res mutant, was significantly disorganized as they showed dilated thylakoids and reduced starch grains (Figure 4b,c). However, after salt treatment, chloroplasts of the RNAi line reorganized their ultrastructure, and the amount of well-stacked grana observed was similar to WT chloroplasts and those of res under same conditions (Figure 4d–f). In conclusion, silencing SLDEAD39 caused changes in the chloroplast ultrastructure that are indistinguishable from changes caused by the res mutation.
3.3 Constitutive expression of SlDEAD39 complements the res mutation
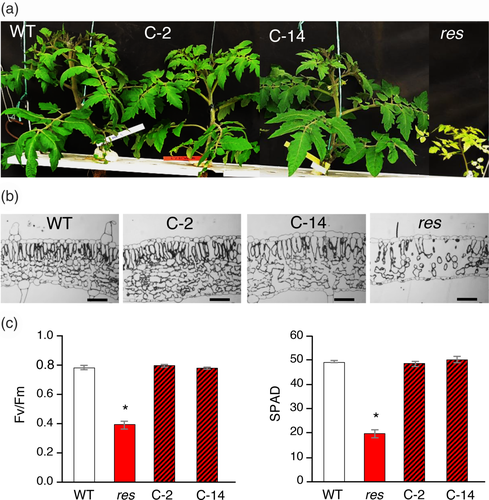
To further corroborate that the res phenotype was due to the mutation in SlDEAD39, a molecular complementation experiment was carried out. With this aim, res mutant explants were transformed with a construct (35S::SlDEAD39) that constitutively expressed the native SlDEAD39 gene and eight independent diploid transgenic lines were obtained (“C” lines in Figure S3). The eight complementation lines showed a WT phenotype indicating that SlDEAD39 complemented the res mutation. Two of these C-lines, namely C-2 and C-14, which showed SLDEAD39 relative expression levels of 13.2- and 5.3-fold, respectively (Figure S3), were selected for a more detailed characterization. The phenotype of both lines was similar to that of WT plants and microscopy analysis transversal leaf sections showed that cell structure abnormalities, which were characteristic of res plants, were not found in both complementation lines (Figure 5a,b). In addition, the absence of leaf chlorosis, the most representative phenotypic trait of res mutant, was confirmed in C-lines by Fv/Fm and SPAD measurements (Figure 5c). Moreover, the development traits analyzed in plants growing under control conditions and salt stress (10 days at 200 mM NaCl), such as number of leaves, stem diameter, number of trusses and axillary buds, as well as the Fv/fm and chlorophyll measures, were very similar in WT and complementation lines, regardless of the growing condition (Figure S5). Taken together, results of the molecular complementation assay confirmed that SlDEAD39 was the gene affected by the res mutation.
We also generated transgenic lines transforming WT plants with the 35S::SlDEAD39 gene construct (OX lines in Figure S3). Seven independent diploid OX lines obtained showed a WT phenotype, and two of them, named OX-4 and OX-9, with SlDEAD39 relative expression levels of 8.6- and 9.0-fold, respectively, were selected for further characterization in control and salt stress conditions (Figure S6). No phenotypic differences were found between overexpression lines and WT plants under either control conditions or salt stress conditions, and plants showed similar values of chlorophyll content, chlorophyll fluorescence, as well as number of leaves, stem diameter, trusses and side branches after salt stress exposition. Altogether, these results indicated that the constitutive expression of SlDEAD39 is able to complement the res mutation and does not change the overall development of wild-type tomato plants.
3.4 The res mutant is defective in chloroplast 23S rRNA processing
In order to elucidate whether SlDEAD39 is involved in the maturation of chloroplast, 23S rRNA, as it was found for the Arabidopsis homologue, AtRH39 (Nishimura et al., 2010), we analyzed the steady state of 23S rRNA fragments by RNA gel blot analysis, using probes covering the different fragments of 23S rRNA sequence (Figure 6a). The 23S rRNA maturation process involves the cleavage of a premature 2.9 Kb sequence at two sites, the hidden break A and B sites (hb-A and hb-B), yielding 0.5, 1.3 and 1.1 Kb mature sequences (Figure 6a). Considering the recovery of a normal phenotype shown by salt-stressed res plants, we analyzed the 23S rRNA processing in the first leaf and stem of mutant and WT plants grown under control conditions and after 5 days of salt stress treatment (5 DST). The 23SFL probe detected a high abundance of immature fragments (2.9 and 2.4 kb) in res plants compared to WT plants, under control conditions, whereas the presence of mature 1.1 and 1.3 kb fragments was almost undetectable in the res mutant leaves and stems (Figure 6b). In addition, the full-length 2.9 kb, the immature fragment of 2.4 kb and the 0.5 kb mature fragment were revealed by the 23SP1 probe in res tissues, unlike the 1.8 kb size intermediate fragment (Figure 6c). The 23SP2 probe detected the 2.9 and 2.4 kb intermediate fragment, but not an appreciable amount of the 1.8 kb intermediate molecules or the mature 1.1 and 1.3 kb fragment (Figure 6d). These results indicated that res mutant is defective in the cleavage in the hidden break site B (hb-B) that generates 1.3 and 1.1 kb mature fragments of 23S rRNA, and, therefore, that SlDEAD39 is likely involved in the chloroplast 23S rRNA maturation of tomato as it was found for AtRH39 in Arabidopsis. Interestingly, the salt-stressed res mutant plants showed an almost normal processing of the chloroplast, 23S rRNA, coinciding with the timing of the phenotypic reversion observed in these plants. Processing of other chloroplast rRNA molecules did not seem to be altered in the res mutant (Figure S7), which would indicate that SlDEAD39 must participate in the processing of the 23S transcript by specifically promoting the cleavage of the immature 23S rRNA at the hidden break B site.
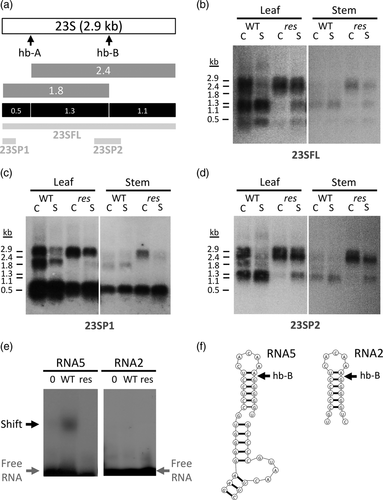
The hypothesis about the molecular function of SlDEAD39 was addressed by protein-RNA binding experiments to assess the ability of recombinant SlDEAD39 to shift RNA molecules that contain the hidden break B. To ascertain the res mutant allele's loss of functionality, the mutant form of the protein was assayed under the same experimental conditions. Results demonstrated that WT SlDEAD39 protein binds to an RNA molecule that contains the hidden break B (hb-B) site, whereas the mutant protein coded by the res mutant allele was incapable of binding to the same RNA fragment (Figure 6e).
3.5 Expression pattern of chloroplast-targeted DEAD-box RHs genes
As described before, res mutant plants restored their phenotype after growing under salt stress conditions. It may be hypothesized that other chloroplast-targeted DEAD-box RH proteins could be induced by salinity and, therefore, complement the loss-of-function of the mutated SlDEAD39. To investigate this, the expression pattern of the nine predicted chloroplast-targeted DEAD-box RHs of tomato, including SlDEAD39, were analyzed in leaves of WT, res mutant and RNAi-9 line plants at day 0 (Control), 1 DST, before phenotype recovery, and 5 DST, when phenotype recovery was evident (Figure 7). The level of SlDEAD39 transcripts was similar in WT and res under control and salt conditions, and much lower in the silencing line as expected. For most of the other SlDEAD-box helicases coding genes (SlDEAD25, SlDEAD28, SlDEAD11, SlDEAD23 and SlDEAD35) none of them are induced by salinity in both res mutant and RNAi-9 line plants, which suggest that these DEAD-box RHs do not seem to be responsible for the recovered phenotype displayed by both res mutant and RNAi plants under salt stress (Figure 7). A different expression pattern was detected for SlDEAD5, SlDEAD17 and SlDEAD33, as they were up-regulated by salinity in the three genotypes analyzed here, that is, WT, res mutant and RNAi-9 plants, although at a distinct level and timing. It is noteworthy to mention that the highest transcript accumulation of these genes was found in the genotypes defective for the SlDEAD39 function.

3.6 The reproductive development of res mutant is increased by salt stress
The previous studies with res mutant have been carried out during vegetative growth of tomato plants at the stage of 4–5 developed leaves, and by applying short- and mid-term salt treatments (Albaladejo et al., 2018; García-Abellán et al., 2015). In this study, we also determine whether the function of SlDEAD39 is needed for tomato development along its whole life-cycle, including the reproductive development phase. For this purpose, adult plants of WT and res mutant were grown in control and 100 mM NaCl for 80 days (80 DST). At the beginning of salt treatment, the res mutant plant showed leaf chlorosis and lower development with respect to WT plants (Figure 8a), the characteristic phenotype of res in control conditions. The evolution of the chlorophyll fluorescence (Fv/Fm) over time showed an increase in res leaves with aging of plants, reflected in a slight re-greening of them. Nevertheless, the increase of chlorophyll fluorescence was more prominent in salt-treated res mutant, which reached similar levels of chlorophyll fluorescence to salt-treated WT plants at the end of the treatment (Figure 8b).
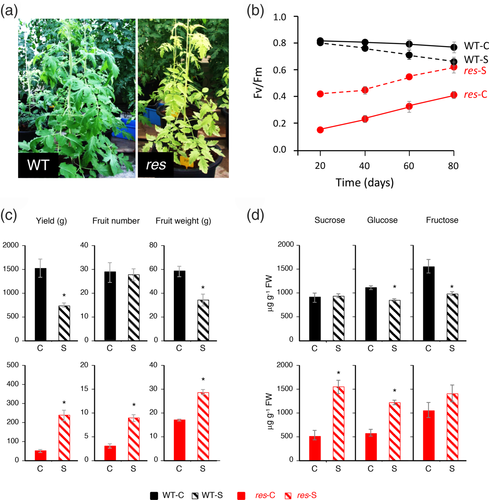
In this assay, we determined that salt stress negatively affected fruit yield due to fruit weight reduction, while the numbers of fruit were similar in control and salt-stressed plants (Figure 8c). According to the lower growth of res plants compared with WT plants, fruit yield was much lower in res than WT plants, under control conditions. However, fruit yield increased significantly in salt-stressed res mutant plants, which showed increased values of both fruit yield components, fruit number and fruit weight (Figure 8c). These results proved that the positive effect induced by salinity on the growth of res mutant occurs not only during the vegetative phase of development, as it had been observed in previous studies, but also during the reproductive phase.
Chloroplasts of adult leaves are the main source of sugars in plants, with sucrose being the main photoassimilate transported from source leaf to sink fruit. In order to elucidate if the lower fruit weight in res mutant, under control conditions, was related to the impaired 23S rRNA processing, and therefore with a defective chloroplast activity, the levels of sucrose, glucose and fructose were analyzed in leaves of WT and res plants at the end of the salt treatment experiment. Under control conditions, leaves of res mutant have significantly lower glucose, fructose and sucrose contents with respect to WT, while the opposite response was observed under salt stress, where sugar contents were significantly higher in salt-treated res leaves compared with WT (Figure 8d). In fact, salinity induced a decrease of glucose and fructose contents in WT leaves, while sucrose and glucose contents were significantly increased in salt-stressed res leaves. This result reflects that the ability of chloroplasts to produce photoassimilates, under control conditions, is impaired in the res mutant, but it is recovered after growing under salt stress conditions.
4 DISCUSSION
4.1 The DEAD-box RNA-helicase SlDEAD39 promotes the proper processing and functionality of chloroplast rRNA
In recent years, several studies in Arabidopsis have reported the essential role of DEAD-box RHs in plant development and stress response (Baruah et al., 2017; Nawaz & Kang, 2017; Nidumukkala et al., 2019). Here, we report the isolation and functional characterization of the tomato, SlDEAD39, gene encoding a DEAD-box RNA-helicase. Tomato plants silencing SlDEAD39 showed a decreased vegetative growth and leaf chlorosis (Figure 2), as well as changes in the ultrastructure of chloroplast (Figure 4), phenotype alterations similar to those previously reported in res mutant plants (García-Abellán et al., 2015). However, two RNAi lines were chlorotic and unable to survive in vitro, whereas another silencing line, RNAi-2, when transferred in vivo failed to accumulate chlorophyll and died after acclimation (Figures 2 and S4). Phenotypic differences between RNAi lines seem to be related to the different levels of SlDEAD39 gene silencing (Figure S3). Our findings are in accordance with other studies reporting chloroplast-localized DEAD-box RH mutants from different species, as most mutants with reduced function of RHs exhibit pale-green seedling phenotypes, whereas complete loss-of-function (null alleles) leads to lethality, as has been described in Arabidopsis rh3, rh22 and rh39 mutants (Chi et al., 2012; Lee et al., 2013; Nishimura et al., 2010). The phenotype of tomato SlDEAD39 RNAi lines also indicated that res is not a complete loss-of-function (null allele) since res mutant plants were able to grow and yield seeded fruits, although they kept the pale-green phenotype and showed a reduced growth rate. In addition, gene expression analyses indicated that SlDEAD39 may exert its function mainly in green tissues and reproductive organs (flowers and fruits), where their transcript levels were very similar, while a lower expression level was detected in root. Constitutive expression of SlDEAD39 in res mutant plants is able to rescue the WT phenotype, both at macroscopic and microscopic levels, as well as the chlorophyll content and the PSII efficiency measured by chlorophyll fluorescence (Figure 5), which together with the phenotype of the RNAi lines, demonstrates that SlDEAD39 is the gene responsible for the res phenotype.
In this research work, we have demonstrated that SlDEAD39 plays a similar role to its Arabidopsis homologue, RH39 (Figure S2), since res mutant is defective in the maturation of 23S rRNA (Figure 6). Thus, SlDEAD39 and RH39 (Nishimura et al., 2010) are both involved in the generation of 1.1 and 1.3 kb rRNA fragments. Furthermore, we have shown that SlDEAD39 specifically participates in the 23S rRNA fragment cleavage located at the hidden break B site (Figure 6). The 23S rRNA is a component of the 50S subunit of chloroplast ribosomes in which the ribosome peptidyl-transferase activity, essential for biosynthesis of plastid proteins, is located. These results are in agreement with the higher expression level of SlDEAD39 gene detected in green tissues where chloroplasts are more abundant (Figure 1c). Several DEAD-box RH coding genes from Arabidopsis have been involved in chloroplast ribosome biogenesis, like RH3, RH22, RH39 and RH50 (Asakura et al., 2012; Chi et al., 2012; Nishimura et al., 2010; Paieri et al., 2018). Tomato chloroplast genome harbors 114 genes (Kahlau, Aspinal, Gray, & Bock, 2006), whereas the size of the plastid proteome is estimated to be around 2,700 proteins (Barsan et al., 2012), which means that more than 95% are nuclear-encoded proteins, synthesized in cytoplasm and transported into the chloroplast to participate in chloroplast function (Pesaresi, Schneider, Kleine, & Leister, 2007). This may be the case for DEAD-box helicase SlDEAD39, whose amino acid sequence contains a predicted N-terminal signal peptide targeting the protein to the plastid (Figure S2). Among the proteins encoded in the tomato plastome, more than 90% are involved in photosynthesis and gene expression (Kahlau et al., 2006). Therefore, the defective maturation of 23S rRNA in res mutant most likely affects the structure and functionality of the chloroplast photosynthetic complex, which is in accordance with the lower photosynthetic efficiency observed in res plants, as well as with their chlorotic phenotype and poor development with respect to WT plants. Taken together, our results prove that SlDEAD39 plays an essential role in 23S rRNA maturation, the major component of the ribosome. Therefore, this RNA helicase protein could be specifically required for upholding the structure and function of the chloroplast through the proper maintenance of ribosome functioning. The lethal phenotype, caused by the SlDEAD39 lack-of-function, supports the functional relevance of this regulatory DEAD-box RH protein during plant development and stress response.
4.2 SlDEAD39 is required for vegetative and reproductive development of tomato
Some studies on DEAD-box RHs have shown evidence that their functions could take place in specific stages of plant development (Cai et al., 2018), as is the case of the Arabidopsis HS3 gene which encodes the plastid DEAD-box RNA helicase 22 (Kanai, Hayashi, Kondo, & Nishimura, 2013). Thus, while 7-day-old hs3-1 mutant seedlings showed a pale green phenotype, and cotyledons and roots were smaller than those of WT, the 14-day-old hs3-1 plants achieved the chlorophyll level of WT and the 30-day-old hs3-1 mutant plants were equal in size to WT plants. Functional characterization of res mutant revealed that SlDEAD39 is crucial in early–mid stages of tomato development, before fructification (Albaladejo et al., 2017; García-Abellán et al., 2015). Thus, we determined that the rate of seed germination of res mutant was lower than WT, the seedlings were pale green and the cotyledons displayed a reduced growth. The chlorotic phenotype and delayed development of res mutant were also described during mid stages (4–5 fully developed leaves) of plant development (García-Abellán et al., 2015). However, an unsolved question was whether this response is maintained throughout the plant's life cycle, including fruit formation. Thus, although slight increases of chlorophyll fluorescence were observed during the aging of res mutant plants, the vegetative growth, as well as the fruit production of mutant adult plants, were significantly lower than WT plants (Figure 8a,c), indicating that chloroplast function is not fully recovered along the life cycle. These results revealed that, unlike RH22 in Arabidopsis (Kanai et al., 2013), SlDEAD39 is needed at all developmental stages of tomato.
Impaired chloroplast function in res adult plants is also supported by the significantly lower amount of the photoassimilates, sucrose, glucose and fructose, observed in mutant leaves compared with WT leaves (Figure 8d). The lower accumulation of photoassimilates in res leaves could be involved in the lower fruit yield of res mutant compared with WT observed under control conditions (Figure 8c). In fact, export of carbohydrates from photosynthetic active leaves provides the substrates required for growth of sink organs such as fruits (Osorio, Ruan, & Fernie, 2014). Therefore, the activity of SlDEAD39 is not only essential during vegetative growth phase of tomato plants, but also significantly influences tomato fruit yield, which suggests that the function of SlDEAD39 is required for tomato development along its whole life cycle.
4.3 Functional redundancy of SlDEAD39 in response to salt stress
Interestingly, salinity reverted the chlorotic phenotype of adult res mutant plants, as was previously observed in early developmental stages (Albaladejo et al., 2017; García-Abellán et al., 2015), and it also improved the capacity of res leaves to produce carbohydrates. Thus, sucrose, glucose and fructose contents were higher in salt-treated mutant leaves than in control mutant ones, reaching even higher levels than those in analogous WT leaves (Figure 8d). The salinity-induced recovery of photosynthetic efficiency and photoassimilates supply could be the cause for the increasing fruit yield in res mutant plants under salt stress condition (Figure 8c).
The recovery from alterations in vegetative growth and fruit production of res mutant, when salt stress was applied, seems to be associated with a similar partial rescue of the correct processing of plastidial 23S rRNA in the mutant plant (Figure 6). In accordance with this observation, transcriptomic analysis of res mutant revealed specific salt- upregulation of key genes involved in photosynthesis, like RUBISCO ACTIVASE1 and ALTERNATIVE OXIDASE1A (Albaladejo et al., 2018), evidencing the enhancement of photosynthetic efficiency under salt stress. On the other hand, some studies have suggested the existence of DEAD-box RHs with similar functions but acting at different developmental stages or environmental conditions (Cai et al., 2018; Pandey et al., 2019; Xu et al., 2013). Therefore, other chloroplast-localized DEAD-box RHs might compensate for SlDEAD39 under salinity conditions. As of today, 42 DEAD-box RHs have been identified in tomato, among which nine are chloroplast-targeted (Pandey et al., 2019; Xu et al., 2013), but their molecular function is still unknown (Cai et al., 2018). Recently, Pandey et al. (2019) showed that heterologous expression of SlDEAD23 and SlDEAD35 genes in yeast enhanced the tolerance of transgenic yeast to salt and cold stresses. In our study, SlDEAD23 was up-regulated in salt-stressed RNAi-9 plants but this gene was not induced in res plants. The opposite expression pattern was observed for SlDEAD35 (Figure 7), which suggests that none of these two genes is responsible for the phenotypic recovery observed in res and SlDEAD39 silencing lines. The only genes upregulated by salinity in WT, res and RNAi plants were SlDEAD5, SlDEAD17 and SlDEAD33, although each showed different induction timing (Figure 7). In addition, the salt-induced expression levels of these genes were higher in res and SlDEAD39 silencing lines, indicating that they, alone or in cooperation, could complement under salinity the function of SlDEAD39 and, therefore, may be responsible for the phenotypic reversion observed in the res mutant.
Molecular analysis showed that SlDEAD5, SlDEAD17 and SlDEAD39 are phylogenetically close, and SlDEAD17 and SLDEAD39 even belong to the same subclass V and form a separate clade in the phylogenetic tree of the protein family (Cai et al., 2018; Pandey et al., 2019). Although SlDEAD33 is phylogenetically distant from SlDEAD39, the SlDEAD33 gene is located in chromosome 10 in a genomic location collinear with the genomic location of chromosome 12 where the SlDEAD39 gene is located, suggesting that both genes were generated by a duplication event within the tomato genome and, therefore, they could be paralogous genes with still partially overlapping functions (Pandey et al., 2019). Our hypothesis about the existence of DEAD-box RHs, functionally redundant with SlDEAD39 under salinity, is supported by the well-documented fact that photosynthesis can function as a stress sensor (Biswal, Joshi, Raval, & Biswal, 2011). Thus, an impaired regulation of genes affecting photosynthesis in chloroplasts, such as occurring in the res mutant, could be sensed, inducing the expression of other nuclear-encoded chloroplast-targeted DEAD-box RHs (SlDEAD5, SlDEAD17, SlDEAD33), which may assist chloroplast gene expression processes under adverse environmental conditions like salinity. Nevertheless, further studies are required to identify proteins or transcription factors that may assist the DEAD-box RHs in mediating their specific functions in stress response.
Taken together, our results seem to indicate that the function of the SlDEAD39 RH is crucial along the whole life cycle of a tomato plant. Thus, the impaired function of SlDEAD39 causes a defective 23S rRNA processing that originates severe alterations in chloroplast structure and function, reducing photosynthesis efficiency and diminishing accumulation of photoassimilates, which, in turn, negatively affects not only plant development but also fruit production. However, under salinity conditions other chloroplast-targeted proteins seem to assist in the 23S chloroplast processing, the DEAD RH proteins SlDEAD5, SlDEAD17 and SlDEAD33 being potential candidates, in order to reestablish the impaired phenotype of SlDEAD39-defective res mutant. Finally, considering the important changes in sucrose and hexoses observed in res mutant leaves, and the crucial role of chloroplasts in the accumulation of photoassimilates in leaves (source organs), the res mutant may be an attractive plant model for source-to-sink relationships studies, a crucial aspect in breeding for fruit yield and even fruit quality.
ACKNOWLEDGEMENTS
This work was supported by the research grants AGL2015-64991-C3-1-R, AGL2015-64991-C3-2-R AGL2015-64991-C3-3-R and AGL2017-88702-C2-1-R from the Spanish Ministry of Economy and Competitiveness (MINECO/FEDER).
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
AUTHOR CONTRIBUTIONS
Carmen Capel, Irene Albaladejo, Isabel L. Massaretto, Fernando J. Yuste-Lisbona, Benito Pineda, Begoña García-Sogo, Trinidad Angosto, Francisco B. Flores and Vicente Moreno contributed to the experiments' design and execution, and the analysis and interpretation of results. Isabel Egea, Rafael Lozano, María C. Bolarín and Juan Capel directed the experiments and wrote the manuscript. All authors have critically reviewed the manuscript.




