An efficient target-mutant screening platform of model variety Ci846 facilitates genetic studies of Setaria
Foxtail millet (Setaria italica), one of the oldest crops originating in China, has increasingly been recognized as a promising C4 model plant due to its compact diploid genome, short growth cycle and self-pollinating nature (Li and Brutnell, 2011). In the past 5 years, significant breakthroughs have been achieved in its basic research and breeding, including high efficient transformation system establishment, telomere-to-telomere (T2T) genome assembly, pan-genome analysis and functional studies (He et al., 2023; Tang et al., 2023; Yang et al., 2020). However, the limited genetic diversity of breeding materials and inefficiencies in identifying target mutants have continued to pose significant challenges in breeding for improved complex agronomic traits and in functional genomics research of this crop.
To broaden novel sources of genetic diversity and enhance mutant identification efficiency for foxtail millet improvement, we constructed a large EMS-mutagenized library and a data-sharing platform based on the model variety Ci846 (www.setariadb.com/millet/mutation, Figure S3), which is a domesticated Setaria variety with well-established transformation system and efficient indoor research platform (Wang et al., 2022). We obtained 9243 M2 lines, of which 775 (8.38%) displayed various morphological variations under field conditions (Figure 1a,b, Table S14 and Figure S4). We sampled 4109 M2 plants and re-sequenced them using a mixed pool strategy, and obtained 1950.76 Gb of pair-end reads from 205 mixed pools, with each pool ranging from 8.6 to 12.2 Gb and an average sequencing depth of 22x (Figure 1c). This represents the largest-scale precise genotype data for mutant libraries in crop species to date. The cleaned short-read sequences were then mapped to the Yugu1-T2T reference genome. The mapping rate and coverage rate of sequencing reads were 98.21 ± 0.37% and 94.24 ± 0.37%, respectively (Table S3). A total of 2,899,449 variations were identified across the 205 mixed pools, with an average mutation density of 1/29.70 Kb across all nine chromosomes (Figure 1d). Each mixed pool contained 1,658,170 ± 11,422 SNPs and 252,784 ± 2402 indels. Among the pools, A173 had the smallest number of variations (1,875,480), while A9 had the largest number (1,989,562) (Table S4). We found that C/G to T/A nucleotide transitions were the most prevalent type (928,081, 38.7%) (Table S7) and the Thr/Val to Ala amino acid substitutions occurred at higher frequency (1.42% and 1.46%) than the average amino acid changed rate (0.16%) (Figure 1e, Table S9). Among all samples, indel size ranged from 1 to 31 bp, with an average of 12 bp, and the most frequent indel type was 1 bp deletions (104,581) (Table S5). A total of 2,368,808 mutations occurred in intergenic regions, followed by 1,850,547 upstream variants, 1,770,535 downstream variants, 413,011 intron variants, 192,335 UTR variants and 248,522 exon variants. Among these exon variants, 128,578 were missense variants, 13,528 were splice-site variants, 15,961 were frameshift deletion/insertion variations and 2851 were stop gain variations (Figure 1e, Table S10). We checked all the 22 genes that have been cloned and reported in foxtail millet to date and found mutations that covered all genes in our mutant library (Table S13). Of these, 14 genes carried missense or frameshift variants in the coding region. To evaluate the accuracy of mutations identified in this study, we randomly selected 73 mutations spanning nine chromosomes for verification by Sanger sequencing (Table S6, Figures S6 and S7). Among them, 93.2% (68/73) mutations were confirmed, indicating the mutations identified in the EMS-induced mutant library are reliable.
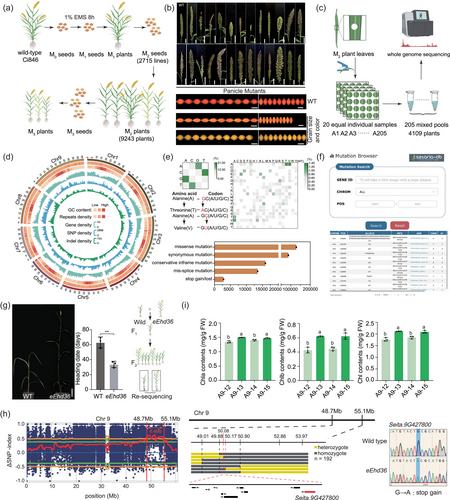
The high-density mutants could be highly beneficial for functional genomics studies as well as breeding progress. To validate the effectiveness of our mutant library in identifying mutants suitable for further functional gene mapping and applications, we identified an extremely early heading mutant, eEhd36, in the M2 population. This mutant exhibited dwarf, slender stems, an extremely early heading date (35 days earlier compared to wild-type Ci846) and showed stable inheritance under field condition from the M2 to M5 generations (Figure 1g). Using BSA-seq and fine mapping on a newly constructed genetic population with the eEhd36 mutant (Table S12), we successfully identified candidate causal gene SiPHYB (Seita.9G427800) within an interval of 50.08–50.17 Mb on Chromosome 9, which has been verified as an important factor affecting the flowering process in plants (Ishikawa et al., 2011), harbours a C-to-T mutation was identified in the second exon of SiPHYB, resulting in a premature termination codon (CAG to TAG) (Figure 1h).
In addition, to further demonstrate the rapid and efficient identification of mutants for target gene functions in our mutant library, we chose SiYGL1 (Seita.9G041600) as an example. SiYGL1 is a yellow-related leaf gene previously reported as regulators of leaf colour and light-use efficiency (Li et al., 2016), and also serves as an assistant selection marker for breeding programs. Using the variation information from the 205 mixed pools, we examined mutation in the SiYGL1 gene and found A9 mixed pool contained SiYGL1 variations (Table S11). We further sequenced each line in the A9 mixed pool by Sanger sequencing at the target variation site. Among the 20 lines of the A9 pool, two mutants, ygl-A9-12 and ygl-A9-14, were identified as carrying homozygous C to T mutations at the target site, resulting in an amino acid change of Val637Ile in SiYGL1 (Figure S5). Then we measured ChIa, ChIb and total chlorphyII contents of two homozygous mutants (ygl-A9-12, ygl-A9-14) and relevant wild-type lines (ygl-A9-13, ygl-A9-15) at the seedling stage. The mutant lines possessed lower chlorophyll contents than the wild-type lines. Specifically, Chla was 1.34 ± 0.038 mg/g FW and 1.41 ± 0.029 mg/g FW in mutants ygl-A9-12 and ygl-A9-14, respectively, compared to 1.496 ± 0.008 mg/g FW and 1.471 ± 0.018 mg/g FW in wild-type lines. Chl b was 0.426 ± 0.040 mg/g FW and 0.446 ± 0.027 mg/g FW in mutants, compared to 0.620 ± 0.007 mg/g FW and 0.621 ± 0.044 mg/g FW in wild-type lines. Total chlorophyll was 1.766 ± 0.077 mg/g FW and 1.852 ± 0.056 mg/g FW in mutants, compared to 2.116 ± 0.015 mg/g FW and 2.092 ± 0.062 mg/g FW in wild-type lines. This indicates that variation in ygl-A9-12 and ygl-A9-14 could be the causal allele responsible for leaf colour change (Figure 1i). These results also suggest that our mutant library is a valuable resource for reverse genetic studies in foxtail millet.
To make target-mutation screening more efficient and feasible, we established an efficient platform for the rapid identification of targeted mutant lines in Setaria by integrating extensive phenotypic and genomic data. All data and tools are publicly accessible on Setaria-DB website (www.setariadb.com/millet/mutation) (Figure 1f). The Ci846 mutant library and whole-genome variation scanning platform provided by this study have established an effective linkage between genomic mutations and agronomic traits in Setaria model variety Ci846, which will serve as an invaluable resource for functional genomics and further accelerate the improvement of foxtail millet, an ancient and globally cultivated cereal crop, in future breeding programs.
Acknowledgements
This study was supported by the National Key Research and Development Program of China (2023YFD1200700/2023YFD1200704), Biological Breeding-Major Projects (2023ZD04076), National Natural Science Foundation of China (32341032, 32301798), The IAEA Coordinated Research Project D24016, China Agricultural Research System (CARS06-14.5-A04), Key Laboratory of Crop Gene Resource and Germplasm Enhancement (MOA), Technology Innovation Program of CAAS and the Youth Innovation Program of Chinese Academy of Agricultural Sciences (Y2024QC01).
Conflicts of interest
The authors declare no conflicts of interest.
Author contributions
G.J., X.D. and Q.H. designed the experiments; H.Z. and H.L. performed the experiments; D.Y., Q.Y., R.Z., L.X., B. Y., L.S., L.Z., H.Z., S.T., L.W., H.W., Y.R., H.Z., Y.Z., E.W., X.M., G.X. and L.Z. provided technical support; H.Z., G.J. and Q.H. wrote the manuscript. All authors read and approved this manuscript.
Open Research
Data availability statement
The data that supports the findings of this study are available in the supplementary material of this article.




