Manipulation of WUSCHEL orthologue expression improves the forage yield and quality in Medicago
Alfalfa (Medicago sativa L.) is a perennial leguminous forage extensively planted around the world (Annicchiarico et al., 2015). As a result, improving alfalfa forage yield and quality is a crucial agricultural goal (Kumar, 2011). Branching traits has a significant impact on the yield of alfalfa (Gou et al., 2018). The previous report showed that HEADLESS (HDL), the orthologue of WUSCHEL (WUS) in M. truncatula, is required for axillary meristem maintenance (Wang et al., 2019), implying HDL has the potential to regulate the number of branches. To test this hypothesis, 35S promoter-driven HDL transgene was introduced into alfalfa. Ten transgenic plants (OX-1, OX-3 and OX-5) with high expression were selected for phenotypic investigation (Figure S1a). Compared with the wild-type, the HDL-OX plants display more branches (Figure 1a–c). Furthermore, overexpressing HDL increases plant height and produces larger, dark green leaves (Figures 1d,e, S1b–f), suggesting that increased HDL activity affects not only branching but also leaf development in alfalfa.
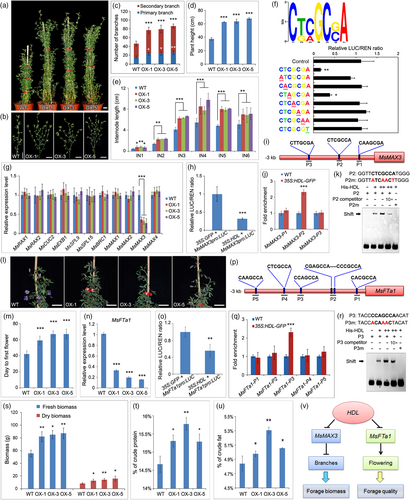
To investigate the regulatory mechanism of HDL, we performed chromatin immunoprecipitation sequencing (ChIP-Seq). In contrast to the conserved WUS-binding motif TAAT, a 7-bp novel HDL binding sequence, CNNGCNA, was identified (Figure 1f). Further analysis showed that HDL interacts with MsTPL/MsTPRs to form a complex and act as a transcriptional repressor (Figures 1f and S2). Point mutation analysis suggested that the core bases are essential for the DNA binding of HDL (Figure 1f). Then, the expression of several known key genes related to branch formation was analysed, and found that the expression of MsMAX3 was significantly reduced in the HDL-OX plants (Figure 1g). MsMAX3 is involved in the biosynthesis of strigolactones (SLs) and localized in both nucleus and cytoplasm (Figure S3a,c). Loss-of-function of MAX3 orthologue mutants exhibited increased shoot branching (Umehara et al., 2008). In addition, down-regulation of the strigolactones receptor MsD14 in alfalfa also leads to increased shoot branching (Ma et al., 2022). Transient expression showed that HDL represses MsMAX3 expression (Figure 1h). The ChIP-qPCR and the electrophoretic mobility shift assay (EMSA) results showed that HDL can directly bind to the P2 fragment of the MsMAX3 promoter (Figure 1i–k). Moreover, down-regulation of MsMAX3 by RNA interference resulted in increased branches (Figure S4), genetically demonstrating that HDL promotes branch formation by inhibiting MsMAX3.
Flowering reduces forage quality to about 45% of the relative feed value (Casler and Vogel, 1999). The HDL-OX plants exhibited a late-flowering phenotype, developing their first flower at a later node than the wild-type (Figures 1l,m; S5). MsFTa1 is localized in the nucleus and cytoplasm (Figure S3b), and mutation of MsFTa1 delays flowering and improves forage quality (Lorenzo et al., 2020; Wolabu et al., 2023). The RT-qPCR analysis showed that MsFTa1 was significantly reduced in HDL-OX plants (Figure 1n). Furthermore, transient expression demonstrated that HDL inhibits MsFTa1 expression (Figure 1o). ChIP-qPCR and EMSA assays revealed that HDL can directly bind to the MsFTa1 promoter (Figures 1p–r), indicating that the down-regulation of MsFTa1 by HDL is responsible for the late-flowering phenotype observed in transgenic alfalfa.”
In addiiton, the HDL-OX plants exhibited a notable increase not only in the leaf/stem ratio, but also in fresh and dry biomass (Figures 1s and S6). The forage quality assay indicated significant increases in the content of crude protein, crude fat, water-soluble sugars, microelements, and neutral and acid detergent fibres in the HDL-OX plants (Figures 1t,u; Table S1). Furthermore, transcriptomic analysis revealed that 5474 genes were differentially expressed in HDL-OX plants, with 3117 genes up-regulated and 2357 genes down-regulated (Figure S7a; Table S2). Among the 18 differential GO terms classified as biological processes, biological regulation, cellular processes, and metabolic processes were the most notable, as they are associated with HDL-OX traits such as increased branching, delayed flowering time, and altered microelement content (Figure S7b; Table S3). Additionally, KEGG analysis showed that these genes were enriched in pathways related to fatty acid biosynthesis, starch and sucrose metabolism, and various amino acid synthesis pathways (Figures S8-S11; Tables S4-S7), supporting the improved forage quality in HDL-OX plants. Despite changes in the expression of many auxin-related genes, such as AUX/IAA, ARF, and SAUR, the auxin content did not significantly change in HDL-OX plants (Figure S12; Table S8). Overall, our study provides evidence supporting the use of HDL as a molecular tool for forage improvement (Figure 1v).
Acknowledgements
We thank Haiyan Yu, Yuyu Guo, and Xiaomin Zhao from the Analysis and Testing Center of SKLMT (State Key Laboratory of Microbial Technology, Shandong University) for assistance with the microscopy.
Conflict of interests
The authors declare no conflict of interest.
Funding
This work was supported by grants from the National Center of Pratacultural Technology Innovation (under preparation) Special Fund for Innovation Platform Construction (CCPTZX2024GJ06), the National Key Research and Development Program of China (2023YFF1001400), the National Natural Science Foundation of China (32170833 and 32201446) and the Natural Science Foundation of Shandong Province (ZR2021QC032 and ZR2022QC109).
Author contributions
H.W. and C.Z. designed the experiments and wrote the paper. H.W., Y.X., Y.W., Z.G. and L.H. performed the experiments. F.Y., S.P.L., S.W.L., Z.L., Y.C., Q.L., C.F., R.L., Q.Y. and Z.W. contributed to the analysis tool and analysed the data.
Copyright transfer statement
This article does not contain any reproduced copyrighted materials from other sources.
Open Research
Data availability statement
The data that support the findings of this study are openly available, further inquiries can be directed to the corresponding author.




