Single-cell transcriptome reveals differentiation between adaxial and abaxial mesophyll cells in Brassica rapa
Mesophyll cells are the main site of photosynthesis and the largest cell population in leaves, with tightly packed cylinder palisade mesophyll cells (PMCs) on the adaxial side and loosely arranged rounded spongy mesophyll cells (SMCs) on the abaxial side. Loss of dorsoventral differentiation of PMCs and SMCs causes alterations in leaf phenotypes (Yu et al., 2020). Brassica rapa encompasses many leafy vegetables with extreme morphological diversity, such as Chinese cabbage with leafy head and pak choi with flat leaves. Exploring the differentiation between PMCs and SMCs and identifying key regulatory genes are important for unravelling the mechanisms underlying leaf development and heading in vegetable crops. However, little is known about them.
Here, we prepared protoplasts from young leaves of Chinese cabbage at the rosette stage for single-cell RNA-seq (scRNA-seq) (Figure 1a). After removing low-quality cells and genes, we obtained 16 055 high-quality cells and 30 214 genes. Our scRNA-seq data showed high reproducibility and a strong correlation with the bulk RNA-seq data (Figure 1b,c). These cells were classified into 17 clusters (Figure 1d). Using the orthologs of marker genes in Arabidopsis, we identified eight cell types, namely mesophyll cells (MCs), epidermis, vasculature, bundle sheath, guard cells, proliferating cells, phloem, and xylem (Figure 1d,e). The expression of Bra001929, a guard cell marker gene, was found to be consistent with the result of in situ RT-PCR (Song et al., 2021). To further verify the annotation result, we compared our results to an Arabidopsis leaf scRNA-seq dataset (Zhang et al., 2021). Pairwise comparisons of cell types and integration analysis of scRNA-seq data between two species both supported our annotation (Figure 1f–h).
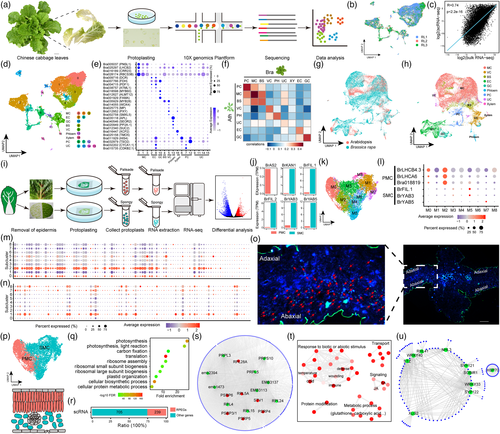
Because MCs were not divided into PMCs and SMCs, we developed an optimized tape-sandwich method to specifically enrich PMCs and SMCs to identify specific marker genes for distinguishing them (Figure 1i). SMCs were enriched according to Uemoto et al. (2018). To collect PMCs, the upper epidermis was removed using tweezers. Then, digestion solution was added to the PMCs and incubated for 30 min to allow the PMCs to be released. We confirmed the successful enrichment of PMCs and SMCs by the specific expression of adaxial and abaxial marker genes (Figure 1j). We identified 6731 differentially expressed genes (DEGs) between them using RNA-seq. Based on many reliable DEGs between adaxial and abaxial domains in Arabidopsis leaves identified by Tian et al. (2019), we selected 433 and 510 potential PMC and SMC marker genes (Table S1), respectively, from overlapping orthologous DEGs shared by B. rapa and Arabidopsis.
To identify PMCs and SMCs, the MC population was re-clustered into 9 subclusters (M0-M8) (Figure 1k). Combining the expression of photosynthetic genes (BrLHCB4.3, BrLHCA6, and Bra018819) and the top 100 potential PMC marker genes, we assigned M0 and M2 to PMC (Figure 1l,m,p). The other 7 subclusters were assigned to SMC by combining the expression of abaxial genes (BrFIL.1, BrYAB3 and BrYAB5) and the top 100 potential SMC marker genes (Figure 1l,n,p). In situ hybridization assays of BrFIL.1 supported the correct definition of SMCs and PMCs (Figure 1o,p).
Using “FindMarkers” in Seurat (v4.0.3), we identified 944 and 1387 genes preferentially expressed in the PMCs and SMCs, respectively (Table S1). Surprisingly, besides identifying many photosynthesis-related genes, a large number of ribosomal protein-encoding genes (RPEGs) (239/944) were enriched in PMCs (Figure 1q,r). Out of the 239 RPEGs found, 27 were orthologous genes of 15 Arabidopsis RPEGs involved in the development of adaxial cells or PMCs (Table S1). Interestingly, protein–protein interaction analysis showed that the top 10 genes with the most connections in PMCs were RPEGs (Figure 1s). These results strongly suggested that RPEGs were involved in PMCs development. Unlike PMCs, SMCs were mainly enriched in the response to abiotic and biotic stimulus, signalling responsive genes (temperature, jasmonic acid, salicylic acid and calcium), and protein modification (Figure 1t). Additionally, almost all the top 10 genes with the most connections in SMCs responded to biotic or abiotic stress (Figure u). Overall, these results indicated that PMCs are the major machine for photosynthesis, while SMCs may be involved in tuning the machine to adapt to external environment.
Surprisingly, we found most of the reported adaxial–abaxial polarity genes to be barely detectable in MCs. To determine why this might be, we collected a series of samples representing the shoot apical meristem (SAM) and different regions of inner and outer leaves at the seedling and rosette stages for RNA-seq (Figure S1a,b). Results showed that most adaxial–abaxial polarity genes were preferentially expressed in the SAM, while they were sharply and continuously downregulated from inner to outer leaves at both stages (Figure S1c). This may be why few adaxial–abaxial polarity genes were detected in MCs, as the middle rosette leaves were collected for scRNA-seq.
Overall, we generated a transcriptome atlas of Chinese cabbage rosette leaves at single-cell resolution. A key finding was the identification of adaxial PMCs and abaxial SMCs from MCs. We identified functional differences between PMCs and SMCs, and the potential role of RPEG in PMC development. Our study also provided many cell type-specific marker genes, which will facilitate the application of scRNA-seq in B. rapa. Comparing the leaf single-cell transcriptome by each cell type, focusing on SMCs and PMCs, across different developmental stages and different subspecies will expand our knowledge of the differentiation of PMCs and SMCs in leaf adaxial–abaxial patterning, and in the processes underlying leaf development and morphogenesis in B. rapa leafy vegetables.
Accession numbers
All sequencing data are available from the NGDC (https://ngdc.cncb.ac.cn/gsa/) under BioProject accession number PRJCA009630.
Acknowledgements
We are grateful to Prof. Xia Cui from CAAS for assistance with protoplast isolation. This work was supported by the National Key Research and Development Program of China (2021YFF1000101), the Central Public-interest Scientific Institution Basal Research Fund (Y2020PT21), and the Agricultural Science and Technology Innovation Program (ASTIP).
Conflict of interest
The authors declare no conflicts of interest.
Author contributions
X.W. and J.W. designed the research. X.G., J.L., and Z.L. prepared materials. X.G., R.L., and Z.Z. performed data analysis. X.G., J.L., and X.W. wrote the manuscript. All authors have approved the manuscript.




