An automated artifact detection and rejection system for body surface gastric mapping
Funding
This work was supported by the New Zealand Health Research Council and the Royal Australasian College of Surgeons' John Mitchell Crouch Fellowship.
Abstract
Background
Body surface gastric mapping (BSGM) is a new clinical tool for gastric motility diagnostics, providing high-resolution data on gastric myoelectrical activity. Artifact contamination was a key challenge to reliable test interpretation in traditional electrogastrography. This study aimed to introduce and validate an automated artifact detection and rejection system for clinical BSGM applications.
Methods
Ten patients with chronic gastric symptoms generated a variety of artifacts according to a standardized protocol (176 recordings) using a commercial BSGM system (Alimetry, New Zealand). An automated artifact detection and rejection algorithm was developed, and its performance was compared with a reference standard comprising consensus labeling by 3 analysis experts, followed by comparison with 6 clinicians (3 untrained and 3 trained in artifact detection). Inter-rater reliability was calculated using Fleiss' kappa.
Key Results
Inter-rater reliability was 0.84 (95% CI:0.77–0.90) among experts, 0.76 (95% CI:0.68–0.83) among untrained clinicians, and 0.71 (95% CI:0.62–0.79) among trained clinicians. The sensitivity and specificity of the algorithm against experts was 96% (95% CI:91%–100%) and 95% (95% CI:90%–99%), respectively, vs 77% (95% CI:68%–85%) and 99% (95% CI:96%–100%) against untrained clinicians, and 97% (95% CI:92%–100%) and 88% (95% CI:82%–94%) against trained clinicians.
Conclusions & Inferences
An automated artifact detection and rejection algorithm was developed showing >95% sensitivity and specificity vs expert markers. This algorithm overcomes an important challenge in the clinical translation of BSGM and is now being routinely implemented in patient test interpretations.
Key points
- Artifact contamination significantly limited the interpretation of traditional electrogastrography data.
- We present an automated algorithm to detect and reject artifacts in body surface gastric mapping recordings.
- The presented algorithm was highly sensitive and specific, and validated for routine clinical use.
1 INTRODUCTION
Chronic gastric symptoms affect up to 10% of adults and children and impose major healthcare and societal cost burden.1, 2 These disorders, which include chronic nausea and vomiting syndromes, gastroparesis, and functional dyspepsia, remain a substantial diagnostic challenge due to overlapping phenotypes and a lack of objective diagnostic tests that can reliably differentiate subgroups to guide individualized therapy.
Body surface gastric mapping (BSGM; also termed high-resolution electrogastrography) is a method for evaluating gastric myoelectrical activity in high spatiotemporal resolution.3-5 These techniques have shown potential to improve accuracy in detecting underlying gastric neuromuscular dysfunction, with studies showing improved correlation of BSGM biomarkers with symptoms compared with gastric emptying or traditional electrogastrography.5-8 A commercial BSGM system has recently become available, offering a standardized clinical test protocol with concurrent symptom capture, followed by spectral and spatial analytics.3, 4, 9
Susceptibility to extrinsic noise and motion artifacts has been a longstanding challenge in the clinical implementation of non-invasive gastrointestinal myoelectrical recordings, posing a known pitfall to previous electrogastrography (EGG) interpretations.4, 7, 10 We were, therefore, motivated to develop a robust and automated artifact detection and rejection algorithm for use in clinical BSGM systems in order to mitigate this problem. To test reliability, the sensitivity and specificity of the new algorithm were validated against a manually labeled reference standard generated by experienced signal processing experts, followed by comparison with gastroenterology clinicians. A visualization system for the new artifact detection method was also designed and implemented, to enable clinical implementation when reporting patient BSGM tests.
2 METHODS
2.1 Study design
This was a prospective validation study of an artifact rejection and detection algorithm, with the code locked prior to participant recruitment. The study was prospectively registered (ClinicalTrials.gov NCT04992884) and managed in accordance with ISO 14155:2020 Clinical investigation of medical devices for human subjects.11 Ethical approval was granted by the Auckland Health Research Ethics Committee (AHREC: AH1130).
2.2 Patient participant protocol
Patient participants with underlying gastric motility disorders underwent BSGM with the Gastric Alimetry™ System (Alimetry, New Zealand), a non-invasive body surface gastric mapping device including a wearable Reader and high-resolution Array (196 cm2, 66 electrodes).3, 6 Patients were ≥18 years, met Rome criteria for a functional gastroduodenal disorder or had a diagnosis of gastroparesis (refer to Appendix S1 for full exclusion criteria). Patients were asked to perform a sequence of artifact-generating activities at 10-min intervals under a uniform protocol including walking, patting the electrode array, resting hands on the array, reading aloud, readjusting seating positions, and coughing for 20 s.
2.3 Artifact detection and rejection algorithm
Raw signal data from the Gastric Alimetry™ system (Alimetry, New Zealand) was processed using the device's proprietary signal processing pipeline, which includes filtering, artifact detection, and signal recovery techniques adapted from those described in detail by Gharibans et al.12 Where data is deemed to be unrecoverable, these periods are marked as artifact and data is not shown on spectral plots (Figure 1), and where data is recoverable, signal processing methods are applied to recover gastric signal data. The post-processed, clean data is then compared with the raw data, for intuitive visualization in clinical outputs (Figure 2).
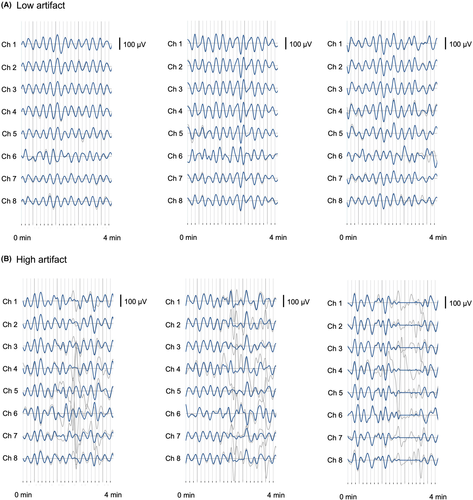
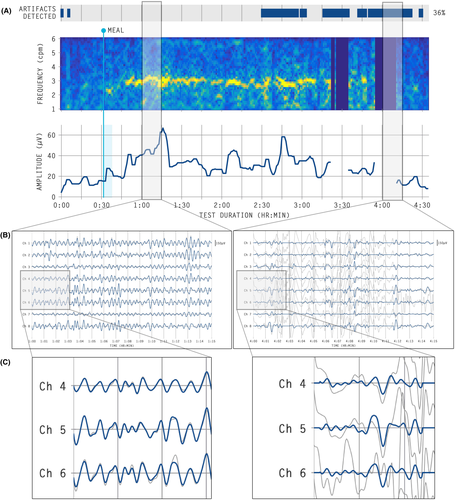
A minimum of 16 images of 4-min segments of data per subject were randomized and sent to clinicians for manual artifact marking (16–18 data periods total). Clinicians and experts were instructed to mark artifacts which we defined as “large spikes in the signal traces” that occurred for >10 s in duration over the entirety of the 4-min window (refer Figure 2).
2.4 Reference standard
The reference standard for assessing algorithm performance was the independent manual marking of artifacts by three expert markers with extensive experience with EGG and signal processing. In addition, real-world performance was assessed by clinicians, with inclusion criteria being specialist gastroenterologists, gastrointestinal surgeons, or advanced clinical trainees in these specialties, of any age and sex. Selected clinicians were consented prior to taking part in the investigation. Three clinicians had no formal training in identifying signal artifacts, and another three independent clinicians received training in artifact identification (refer Appendix S1). Consensus between markers (defined as agreement of ≥2 markers) was used to establish the reference standard for each group.
2.5 Statistical analysis
Fleiss' kappa statistic was evaluated to determine inter-rater reliability.13 Bootstrapping was used to calculate 95% confidence intervals for Fleiss' kappa statistic, sensitivities, and specificities. Further analysis details can be found in the Appendix S1.
3 RESULTS
Ten female participants meeting Rome IV criteria for functional dyspepsia or chronic unexplained nausea and vomiting, of average age 40 (standard deviation 19.7) and average body mass index 25.0 (SD 4.8) provided data (refer Appendix S1; Tables S1 and S2).
3.1 Algorithm comparison with expert markers
Fleiss' Kappa statistic was 0.84 (95% CI: 0.77–0.90) demonstrating “almost perfect agreement” between expert markers. Compared with gold-standard manual artifact marking by experts, of a total of 176 samples, the algorithm correctly classified 168 samples, giving an overall accuracy of 95%. The sensitivity and specificity of the automated algorithm were 96% (95% CI: 91%–100%) and 95% (95% CI: 90%–99%), respectively (Tables S3 and S4).
3.2 Algorithm comparison with untrained clinicians
Fleiss' Kappa statistic was 0.76 (95% CI: 0.68–0.83) demonstrating “substantial agreement” between raters. Of a total of 176 samples, the algorithm correctly classified 152 samples, giving an overall accuracy of 86%. The sensitivity and specificity of the automated algorithm were 77% (95% CI: 68%–85%) and 99% (95% CI: 96%–100%), respectively (Tables S3 and S4).
3.3 Algorithm comparison with trained clinicians
Fleiss' Kappa statistic was 0.71 (95% CI: 0.62–0.79) demonstrating “substantial agreement” between raters. Of a total of 176 samples, the algorithm correctly classified 161 samples, giving an overall accuracy of 91%. The sensitivity and specificity of the automated algorithm were 97% (95% CI: 92%–100%) and 88% (95% CI: 82%–94%), respectively (Tables S3 and S4).
Sensitivity and specificity of labelers compared with consensus can be found in Tables S5 and S6.
4 DISCUSSION
This study introduces an automated artifact detection and rejection algorithm for BSGM, showing high sensitivity and specificity against a manually marked reference standard provided by signal processing experts. The algorithm is demonstrated to present a valuable practical step in the translation of BSGM, by enabling easy and effective identification, rejection, and visualization of artifacts in clinical applications.
Cutaneous gastric myoelectrical signals are approximately 100× weaker than cardiac signals, and diminish exponentially with distance from the source.14 Contamination of these weak signals with extrinsic artifacts, which may be difficult to reliably discriminate from signal, was one important factor limiting the utility of traditional EGG.7 Standard EGG analytics presented no convenient solution to this issue, which can be further exacerbated by the analytical technique of binning data into several-minute windows leading to segments of data loss. While some previous EGG artifact reduction methods did attempt to overcome these issues, the approaches were limited to research settings, did not offer intuitive visualization to users, and were not validated against manual markers.15, 16 The new method presented here is shown to be validated using patient data and is embedded within a commercial system, ensuring it will be available to clinical users when interpreting patient data.
The new algorithm rejects artifacts while automatically recovering the underlying data when possible, thereby maximizing obtained continuous signal data.15 Such approaches have shown potential to achieve improved motility correlations with antroduodenal manometry, compared with simply excluding periods with artifact completely.12 In addition, continuous high-resolution spectral data is anticipated to be valuable for characterizing meal response time courses and enabling symptom correlations.3, 6
Recently presented BSGM systems generate vast volumes of data from 64 electrodes over up to 4.5 h of recordings,3 compared with traditional 3–6 electrode EGG systems applied for up to 90 minutes. This scale of data generation renders manual artifact identification impractical, such that robust automated techniques are not only desirable but important for clinical utility. Automated artifact rejection also enhances ease of use for clinicians.
One of the challenges in developing this algorithm was determining an accurate gold standard to the presence of artifacts in EGG signals. The current standard practice is manual marking of artifacts, as previously applied extensively in serosal slow-wave analytics.14, 17 This process was optimized by utilizing a consensus of experienced signal processing experts who independently assessed randomized segments of data. However, the clinical evaluators in this study with limited experience in myoelectrical signal analytics tended to show over-marking of trivial signal deviations, as evidenced by reduced sensitivities and higher inter-rater variability. This effect was effectively overcome by developing a training platform to provide a more accurate standard to which an algorithm could be compared with clinicians, with improved results.
In summary, the automated artifact rejection algorithm presented here has been confirmed to accurately detect artifacts and is validated by comparison with both experts and end-clinician users in identifying artifacts, presenting a valid and convenient approach to managing artifacts in BSGM data. The new technique has now been made available for clinical use in patient BSGM test interpretations.
CONFLICT OF INTEREST
AG, PD, CNA, GOG hold grants and intellectual property in the field of GI electrophysiology and are members of University of Auckland spin-out companies: The Insides Company (GO), FlexiMap (PD), and Alimetry (AG, SC, SW, JSTW, PD, CNA and GOG). CV has no relevant conflicts to declare.
5 AUTHOR CONTRIBUTIONS
SC, SW, PD, GOG, AG, were involved in study conception and design. SC, SW, GS, JST, AG, were involved in data collection. SC, GS, CV, SW, GS, JST, PD, were involved in the data analysis and interpretation. SC, GS, CV, GOG, AG, CAN, PD were involved in drafting the manuscript. SC, GS, CV, SW, GS, JST, PD, CAN, GOG, AG were involved in critical revisions/final approval of the manuscript.
ACKNOWLEDGEMENTS
Open access publishing facilitated by The University of Auckland, as part of the Wiley - The University of Auckland agreement via the Council of Australian University Librarians.




