The highly diverged trypanosomal MICOS complex is organized in a nonessential integral membrane and an essential peripheral module
Summary
The mitochondrial contact site and cristae organization system (MICOS) mediates the formation of cristae, invaginations in the mitochondrial inner membrane. The highly diverged MICOS complex of the parasitic protist Trypanosoma brucei consists of nine subunits. Except for two Mic10-like and a Mic60-like protein, all subunits are specific for kinetoplastids. Here, we determined on a proteome-wide scale how ablation of individual MICOS subunits affects the levels of the other subunits. The results reveal co-regulation of TbMic10-1, TbMic10-2, TbMic16 and TbMic60, suggesting that these nonessential, integral inner membrane proteins form an interdependent network. Moreover, the ablation of TbMic34 and TbMic32 reveals another network consisting of the essential, intermembrane space-localized TbMic20, TbMic32, TbMic34 and TbMic40, all of which are peripherally associated with the inner membrane. The downregulation of TbMic20, TbMic32 and TbMic34 also interferes with mitochondrial protein import and reduces the size of the TbMic10-containing complexes. Thus, the diverged MICOS of trypanosomes contains two subcomplexes: a nonessential membrane-integrated one, organized around the conserved Mic10 and Mic60, that mediates cristae formation, and an essential membrane-peripheral one consisting of four kinetoplastid-specific subunits, that is required for import of intermembrane space proteins.
Graphical Abstract
Introduction
All mitochondria capable of oxidative phosphorylation contain cristae, invaginations which enlarge the area of the inner membrane (IM). Cristae divide the mitochondrial IM into two domains: the inner boundary membrane, which is adjacent to the outer membrane (OM) and that is enriched for protein import complexes, and the cristae membranes, where the electron transport chain complexes and the ATP synthase are localized. The two membrane domains are separated by the cristae junctions (CJs), narrow neck-like structures in which the IM is sharply bent. CJs are formed by the mitochondrial contact site and cristae organizing system (MICOS), a hetero-oligomeric protein complex consisting of at least 6–7 subunits in yeast and mammals, respectively (Harner et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011; Alkhaja et al., 2012; Kozjak-Pavlovic, 2017). Except for the one or two peripheral membrane proteins that face the intermembrane space (IMS) all are integral IM proteins. MICOS is essential for the building of CJs and contact sites to the OM and thus for the formation of submitochondrial microcompartments optimized for oxidative phosphorylation (van der Laan et al., 2016; Rampelt et al., 2017; Kozjak-Pavlovic, 2017; Schorr and van der Laan, 2018; Quintana-Cabrera et al., 2018). Moreover, the contact sites were shown to promote mitochondrial protein import (von der Malsburg et al., 2011; Bohnert et al., 2012; Wrobel et al., 2015) and facilitate lipid transport (Harner et al., 2014; Aaltonen et al., 2016; Michaud et al., 2016). In a wider context, MICOS can be viewed as a mitochondrial IM organizing structure that directly or indirectly influences many mitochondrial features including calcium uptake (Schorr and van der Laan, 2018) and mitochondrial DNA organization (Itoh et al., 2013; Li et al., 2016). It is therefore no surprise that MICOS dysfunction has been linked to a number of human diseases (van der Laan et al., 2016; Rampelt et al., 2017; Kozjak-Pavlovic, 2017; Wollweber et al., 2017).
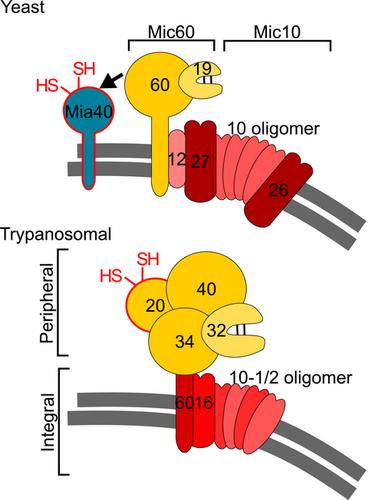
Studies in yeast and humans identified two distinct MICOS subcomplexes organized around each of its two core subunits Mic10 and Mic60 respectively (Bohnert et al., 2015; Ding et al., 2015; Friedman et al., 2015; Guarani et al., 2015; Ott et al., 2015; Li et al., 2016). The yeast Mic60 subcomplex contains Mic60 and Mic19 and mediates contact site formation by interaction with various OM proteins and protein complexes including the sorting and assembly machinery (SAM), the protein translocase of the OM (TOM) as well as the voltage dependent anion channel (VDAC) (Korner et al., 2012; Zerbes et al., 2012; Schorr and van der Laan, 2018). The Mic10 subcomplex on the other hand consists of Mic10, Mic12, Mic26 and Mic27 and appears to be mainly devoted to the formation and maintenance of CJs (Zerbes et al., 2012).
The MICOS complex has experimentally been studied mainly in yeast and humans, both of which belong to the same eukaryotic supergroup Opisthokonta (Burki, 2014; Huynen et al., 2016). However, two MICOS subunits, Mic10 and Mic60, are evolutionarily conserved in all eukaryotes that have cristae-containing mitochondria (Munoz-Gomez et al., 2015; 2017). Moreover, Mic60 orthologues are found in α-proteobacteria, suggesting that a Mic60-like protein was present in the bacterial ancestor that was converted into the mitochondrion (Munoz-Gomez et al., 2015; 2017; Tarasenko et al., 2017).
The only experimentally studied MICOS complex outside the Opisthokonta is the one of the parasitic protist Trypanosoma brucei, a member of the eukaryotic supergroup Excavata (Burki, 2014; Munoz-Gomez et al., 2015). Identification of the Mic10 orthologues in the T. brucei genome opened the way for the characterization of its MICOS complex (Kaurov et al., 2018). Pulldown experiments using epitope tagged Mic10 orthologues recovered 9 proteins. Moreover, the same set of proteins was consistently detected by several reciprocal immunoprecipitations (IPs) using the newly identified MICOS subunits. Three of them are conserved: the two Mic10 paralogues, termed TbMic10-1 and TbMic10-2, and a putative Mic60 orthologue, termed TbMic60, which shows a similar domain structure as the yeast protein but lacks the mitofilin domain. It has been suggested that the absence of this domain might be compensated for by association of TbMic60 with the peripheral membrane protein TbMic34 which has two coiled–coiled domains (Kaurov et al., 2018). The remaining trypanosomal MICOS subunits TbMic16, TbMic17, TbMic20, TbMic32, TbMic34 and TbMic40 are unique to the kinetoplastid flagellates. The T. brucei MICOS was shown to be required for the maintenance of the typical discoidal cristae found in the kinetoplastids and euglenids. Moreover, ablation of the thioredoxin-like protein TbMic20 inhibited import of IMS-localized proteins suggesting it may be the long sought functional analogue of Mia40 (Kaurov et al., 2018), that seems to be absent from kinetoplastids (Basu et al., 2013; Haindrich et al., 2017). Mia40 is the central catalyst of mitochondrial intermembrane space assembly (MIA) pathway that mediates the oxidative-folding IMS proteins to sequester them in the IMS (Stojanovski et al., 2012; Mordas and Tokatlidis, 2015).
All trypanosomal MICOS subunits are present in a complex bigger than 1 MDa that was difficult to resolve by blue native polyacrylamide electrophoresis (BN-PAGE) (Kaurov et al., 2018). However, using IPs and BN-PAGE it was not possible to deduce whether the trypanosomal MICOS complex is organized into two subcomplexes as is the case in yeast and mammals. Thus, we decided to combine knockout/knockdown of individual MICOS components with proteome-wide analyses to investigate which subunits of the trypanosomal MICOS subunits might form networks whose members depend on each other.
Results
ΔTbMic10 strains define a network of integral membrane MICOS subunits
Using serial transfections two knockout cell lines, ΔTbMic10-1 and ΔTbMic10-2, were produced in which both alleles of either TbMic10-1 or TbMic10-2 were replaced by antibiotic resistance cassettes. Growth of the ΔTbMic10-2 cell line was not altered and that of the ΔTbMic10-1 cell line was only slightly affected in media containing or lacking glucose (Fig. 1). In the latter case, cells depend on oxidative phosphorylation for energy conversion (Lamour et al., 2005). Thus, as reported before (Kaurov et al., 2018), neither TbMic10-1 nor TbMic10-2 is essential for growth of procyclic T. brucei in culture.
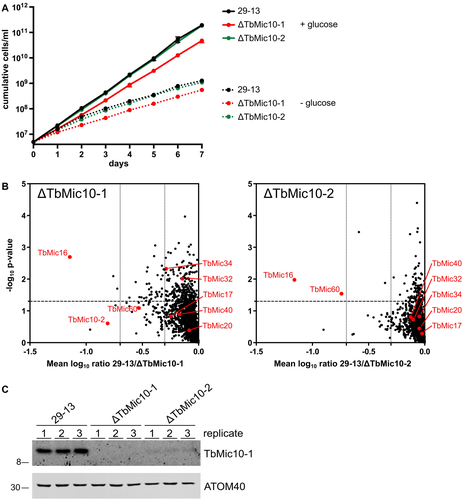
Growth and proteomic characterization of the ΔMic10-1 and ΔMic10-2 cell lines.
A. Growth curves of the ΔMic10-1, ΔMic10-2 and the corresponding parent cell line T. brucei 29-13 in SDM80 medium containing or lacking glucose.
B. Graph on the left indicates proteins reduced in abundance in mitochondria-enriched fractions of the ΔMic10-1 cell line. MS-based quantification of proteins from the ΔMic10-1 cell line versus T. brucei 29-13 cells was based on peptide stable isotope dimethyl labeling (n = 3). Vertical dashed lines indicate a two- and fivefold reduction respectively. Horizontal dashed line indicates a p-value of 0.05. MICOS subunits are indicated with red dots. Graph on the right shows the same analysis for the ΔMic10-2 cell line. For a complete list of protein see Table S1.
C. Immunoblots of crude mitochondrial extracts from T. brucei 29-13, ΔMic10-1 and ΔMic10-2 grown in standard medium were probed for TbMic10-1 and ATOM40 as a loading control.
Next, we compared the proteomes of mitochondria-enriched fractions of the ΔTbMic10-1 cell line with its parental cell line having two intact alleles of TbMic10-1. Proteins in the fractions of the two cell lines were quantified by mass spectrometry (MS) using peptide stable isotope dimethyl labeling (Peikert et al., 2017). Lack of TbMic10-1 resulted in a significant, approximately 16-fold, reduction of TbMic16 (Fig. 1B, left panel). TbMic10-2 and TbMic60 are also downregulated although to a lesser extent and with quite high variability between sample replicates. All other MICOS subunits remained unaltered or were only marginally downregulated (Fig. 1B, left panel).
The corresponding experiment for the ΔTbMic10-2 cell line gave comparable results (Fig. 1B, right panel). Again, an approximately 16-fold decrease of TbMic16 was observed. Moreover, the level of TbMic60 was significantly reduced even to a higher extent as in ΔTbMic10-1. Like in the ΔTbMic10-1 cell line, the levels of all other detected MICOS subunits were not affected. Interestingly, TbMic10-1 could not be detected by MS. Immunoblot analysis (Fig. 1C) showed that knockout of TbMic10-2 causes a dramatic reduction in TbMic10-1 steady-state levels. In summary, these results demonstrate that TbMic10-1, TbMic10-2, TbMic60 and TbMic16, all of which are integral membrane proteins, form an interdependent network. Ablation of either of the two paralogous MICOS core subunits, TbMic10-1 and TbMic10-2, impairs the stability of all other members of this network.
To investigate whether the TbMic10-containing network is required for cristae formation or maintenance, we analyzed ΔTbMic10-1 and its parental cell line by transmission electron microscopy (EM). The ratio between the lengths of the cristae membranes and the OM (a proxy for the adjacent inner boundary membranes) was blindly quantified from a randomized selection of pictures from both cell lines. A significant reduction of cristae membranes in the ΔTbMic10-1 cell line was evident, suggesting that the TbMic10-containing network is required for CJ formation (Fig. 2). Moreover, it can be concluded that cristae membranes can be dramatically reduced without affecting growth of procyclic T. brucei in culture. This is further supported by the observation that in the T. brucei strain 29-13, investigated in the present study, ablation of TbMic60 does not interfere with growth (Fig. S1).
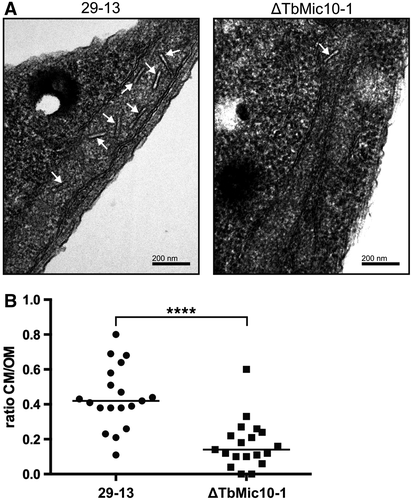
TbMic10-1 is required for cristae formation.
A. Transmission EM images from T. brucei 29-13 (left) and the ΔTbMic10-1 cell line (right). The white arrows mark the cristae.
B. Quantitation of the ratio between the lengths of all discernable cristae membranes and discernable outer membranes in individual transmission EM images of the T. brucei 29-13 (n = 19) and the ΔTbMic10-1 cell lines (n = 19). **** indicates P < 0.0001.
Knockdown of TbMic34 defines a soluble second MICOS subunits network
TbMic34 is a trypanosomal MICOS subunit that is not part of the TbMic10-containing network defined above. The protein is of special interest since it was suggested to compensate for the lacking mitofilin domain of TbMic60 (Kaurov et al., 2018). Carbonate extraction at high pH shows that C-terminally tagged TbMic34 fractionates with soluble proteins (Kaurov et al., 2018) indicating that tagged TbMic34 is a peripheral membrane protein that is tightly associated with the MICOS complex.
Knockdown of TbMic34 causes a growth arrest (Fig. 3A). However, the levels of TbMic10-1, the atypical protein translocase of the OM 40 (ATOM40) (Schneider, 2018) and VDAC remain constant (Fig. 3B). Interestingly, we observe an accumulation of cytochrome oxidase subunit 4 (CoxIV) uncleaved precursor and in parallel a reduction of its mature form, which indicates an inhibition of mitochondrial protein import (Wenger et al., 2017) (Fig. 3B, lowest panel). A protein import phenotype related to CoxIV was also evident after knockdown of TbMic20 (Fig. S2), the putative catalyst for import of IMS proteins (Kaurov et al., 2018) such as the small Tim proteins (Wenger et al., 2017). Inhibition of protein import after knock down of TbMic20 was also directly analyzed using in vitro assays. Fig. S2C shows that when in vitro translated CoxIV precursor is incubated with mitochondria that were isolated from the uninduced TbMic20-RNAi cell line the substrate is processed to its mature form and becomes resistant to externally added proteases. Moreover, import as expected for an IM protein, requires an intact membrane potential. However, when the same experiment was done with mitochondria isolated from the induced TbMic20 RNAi cell line essentially no import was observed, irrespectively whether they had an intact membrane potential or not.
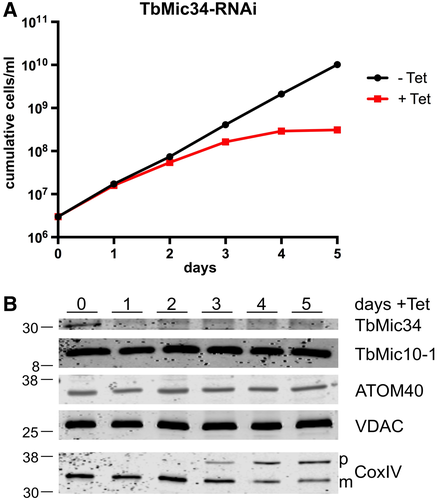
Ablation of TbMic34 does not affect TbMic10-1 levels but inhibits protein import.
A. Growth curves of uninduced (-Tet) and induced (+Tet) TbMic34 RNAi cell lines.
B. Immunoblots of total cellular extracts from the TbMic34 RNAi cell lines induced for the indicated days were probed for TbMic34, TbMic10-1, ATOM40, VDAC and CoxIV using polyclonal antisera. The positions of precursor and mature form of CoxIV are indicated.
To get a global view of the effect upon TbMic34 knockdown, we analyzed the proteome of mitochondria-enriched fractions using SILAC at two time points after RNAi induction. 2.5 days after RNAi induction a downregulation of a subset of MICOS subunits was observed (Fig. 4A). Besides the RNAi target TbMic34, the levels of the essential MICOS components TbMic20, TbMic32 and TbMic40 were significantly reduced, whereas the TbMic10-containing network was not affected. This finding defines a second network of MICOS subunits whose stability depends on TbMic34. All of its members are membrane-associated soluble proteins that reside in the IMS.
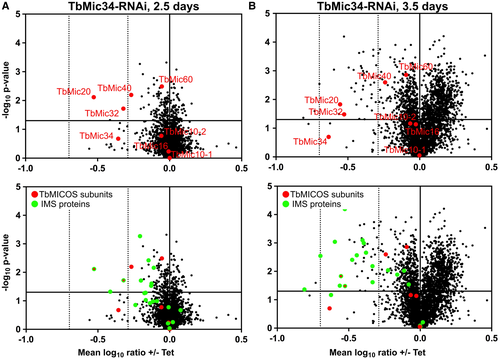
At the later RNAi induction time point (3.5 days) indirect effects could be observed (Fig. 4B). Besides the downregulation of the TbMic34-containing network, a reduction in the levels of many proteins that are known or predicted to be localized, in the IMS (Peikert et al., 2017) was measured. Thus, the observed results phenocopy what has previously been observed for the TbMic20 knockdown cell line (Kaurov et al., 2018) and therefore support the notion that TbMic20 might be a functional analogue of Mia40 which has not been identified in trypanosomatid genomes so far (Basu et al., 2013).
Moreover, many of the other proteins that are downregulated at the later time point are subunits of respiratory complexes (Table S2). A likely cause for this could be the reduced abundance of the IMS-localized small Tim chaperones (Wenger et al., 2017) which are required for the import of TbTim17, the core subunit of the trypanosomal TIM complex (Singha et al., 2008; Harsman et al., 2016).
Characterization of TbMic32 confirms a second MICOS network
To further address the possible existence of a second MICOS network comprised of soluble IM proteins, we characterized another of its subunits, TbMic32. Fig. 5A shows that a C-terminally tagged version of TbMic32 is recovered in a crude mitochondrial fraction. Moreover, after carbonate extraction at high pH the tagged TbMic32 is found in the supernatant indicating that it is a soluble protein (Fig. 5A). TbMic32 contains two CX9C motifs near its C-terminus, a signature found in many IMS-localized proteins (Stojanovski et al., 2012; Mordas and Tokatlidis, 2015). In line with this, the protein gets depleted after RNAi-mediated ablation of TbErv1, an essential import factor for IMS proteins (Fig. 5B) (Peikert et al., 2017).
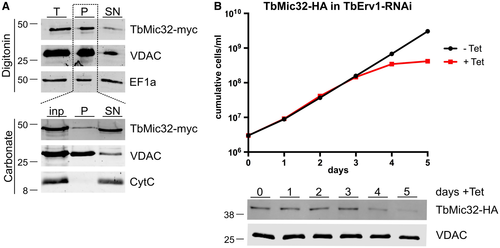
TbMic32 is a soluble IMS-localized protein.
A. Top panels, digitonin extraction of total cells (T) to separate a mitochondria-enriched fraction (P) from the cytosol-containing supernatant (SN). The tagged TbMic32 co-fractionates with the mitochondrial marker VDAC while the cytosolic elongation factor 1A (EF1a) stays in the supernatant. Bottom panels, carbonate extraction at pH 11.5 of the mitochondria-enriched pellet fraction (inp) probed for TbMic32-myc. The pellet (P) and the supernatant (SN) fractions correspond to integral membrane and soluble proteins. VDAC and cytochrome C (Cyt C) serve as markers for integral and peripheral membrane proteins respectively.
B. Growth curve of an uninduced and induced TbErv1-RNAi cell line that constitutively expresses C-terminally HA-tagged TbMic32. Bottom panels, immunoblots of total cellular extracts of the same cell line probed for HA-tagged TbMic32. Time of induction is indicated. The OM protein VDAC serves as loading control. [Colour figure can be viewed at wileyonlinelibrary.com]
RNAi-mediated downregulation of TbMic32 results in a growth arrest as previously reported (Fig. 6A) (Kaurov et al., 2018). Moreover, concomitant with its ablation, a reduction of TbMic34 is observed, whereas the levels of TbMic10-1 and two mitochondrial OM membrane proteins (VDAC and ATOM40) are not affected (Fig. 6B). Interestingly, a reduction of mature CoxIV and an accumulation of its precursor was observed, mirroring the mitochondrial protein import defect seen upon TbMic34 knockdown. The levels of ATOM40 (Schneider, 2018) and VDAC were unaffected under these conditions (Fig. 6B; lower panels). Thus, TbMic32 affects the abundance of TbMic34 but not of TbMic10-1, leading to the conclusion that these two proteins are members of the second network of interdependent MICOS subunits.
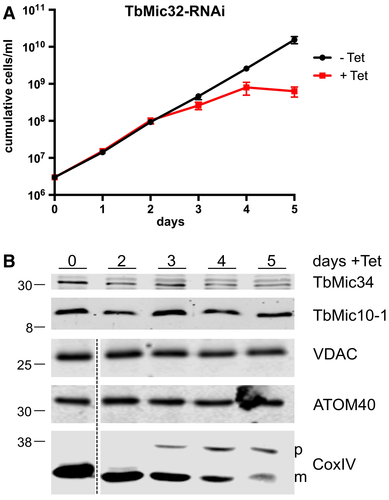
Ablation of TbMic32 affects TbMic34 but not TbMic10 levels and inhibits protein import.
A. Growth curve of an uninduced and induced TbMic32-RNAi cell line.
B. Immunoblots of total cellular extracts from the TbMic34 RNAi cell lines induced for the indicated days were probed for TbMic34 and TbMic10-1 (top panel) or VDAC, ATOM40, and CoxIV (bottom panel) using polyclonal antisera. The positions of precursor and mature form of CoxIV are indicated.
MICOS subunit networks reflect MICOS subcomplexes
Using proteome-wide analyses of knockdown/knockout cell lines for selected MICOS subunits, we have defined two nonoverlapping networks of MICOS subunits. The first one consists of integral membrane proteins, while the other one is composed of soluble IMS-localized proteins. It is difficult to prove that the two networks correspond to physical subcomplexes because IPs of individual subunits always recover all 9 trypanosomal MICOS subunits. Moreover, BN-PAGE analyses show that all tagged MICOS subunits are present in a difficult-to-resolve, very large complex of more than 1 MDa with little evidence for subcomplexes (Kaurov et al., 2018). A caveat is that most of the available data have been obtained with tagged proteins, which may not be as efficiently integrated into the MICOS complex or subcomplexes as the endogenous untagged proteins. Using antibodies specific for TbMic10-1 allowed, the analysis of TbMic10-1-containing complexes after the ablation of other MICOS subunits (Fig. 7). In untreated cells, most TbMic10-1 is found in the high-molecular weight MICOS complex that was shown to contain all tagged MICOS subunits. In addition, two low abundance TbMic10-1-containing subcomplexes of approximately 80 and 120 kDa could be detected. Knockdown of TbMic32, TbMic34 or TbMic20, all members of the second, soluble network, resulted in essentially the same phenotype: a partial shift of the high molecular weight MICOS complex to three low-molecular weight Mic10-1-containing subcomplexes of approximatey 80, 120 and 200 kDa. Their existence suggests that deficiency of the second MICOS network allows the TbMic10-1-containing network to be recovered as smaller physical subcomplexes. The molecular weight differences between these subcomplexes may be due to different amounts of TbMic10-1, and possibly TbMic10-2, as their orthologues in yeast and mammals are known to form oligomers (Barbot et al., 2015; Bohnert et al., 2015).
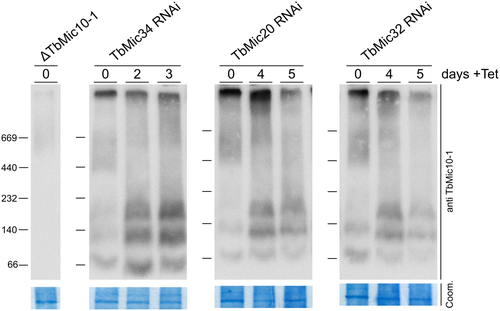
Discussion
BN-PAGE shows that each of the 9 tagged MICOS subunits comigrates in a very large complex of more than 1 MDa (Kaurov et al., 2018). Using proteome-wide analysis of cell lines depleted for specific subunits we show that in trypanosomes this high-molecular weight MICOS complex consists of two largely independent functional networks of MICOS subunits that likely represent physical subcomplexes.
The first network consists of the integral membrane proteins TbMic10-1, TbMic10-2, TbMic16 and TbMic60, whereas the second network comprises the soluble IMS-localized proteins TbMic20, TbMic32, TbMic34 and TbMic40. TbMic17 was not downregulated in the proteomes of the ΔMic10-1 and ΔMic10-2 cell lines and is therefore not part of the first network. Whether it contributes to the second one is unclear since it was not detected in the SILAC RNAi analysis of TbMic34 RNAi.
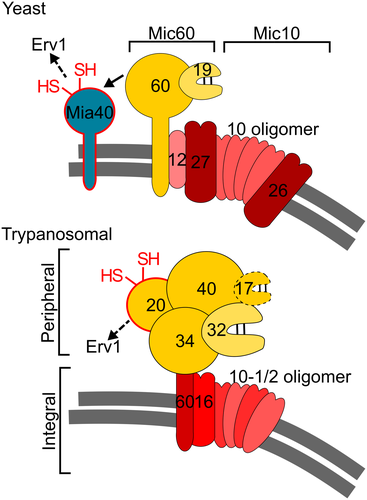
The bipartite nature of the MICOS complex is conserved among trypanosomes, yeast and humans. However, the resulting subcomplexes are very different in opisthokonts as compared to trypanosomes. In yeast and humans they are each organized around one of the widely conserved core components, the integral membrane proteins Mic10 and Mic60 respectively. Mic10 is associated with the integral membrane proteins Mic26 and Mic27 forming the Mic10 subcomplex, whereas the Mic60 subcomplex includes the peripheral membrane proteins Mic19 in yeast or Mic25, the paralogue thereof in humans. The main function of the Mic10 subcomplex appears to be membrane sculpting and thus the building of the CJs, whereas the Mic60 subcomplex mainly forms the contact sites between the OM and the IM, although Mic60 also has a membrane-bending activity (Hessenberger et al., 2017; Tarasenko et al., 2017). Both subcomplexes are independently anchored in the cristae membranes, as most of their subunits are integral membrane proteins. Finally, it was shown that the integral IM protein Mic12 in yeast, or its orthologue in humans QIL12/MIC13 (Guarani et al., 2015), mediate the lateral interaction of the two subcomplexes (Bohnert et al., 2015; Ding et al., 2015; Friedman et al., 2015; Guarani et al., 2015; Ott et al., 2015; Li et al., 2016).
While the trypanosomal MICOS contains orthologues of Mic10 and Mic60, the way the complex is organized in two networks is strikingly different from what is observed in yeast and mammals. One network of MICOS subunits is composed of the four integral membrane proteins TbMic10-1, TbMic10-2, TbMic16 and TbMic60. Thus, unlike in opisthokonts, both conserved MICOS subunits, the Mic-10 paralogues TbMic10-1 and TbMic10-2 together with TbMic60 belong to the same network, whereas all soluble IMS-localized proteins – TbMic34, TbMic32, TbMic20 and TbMic40 – comprise the second network. Therefore, rather than the lateral association of the two membrane-embedded MICOS subcomplexes that is described in opisthokonts, a vertical association of the trypanosomal subcomplexes between a module consisting exclusively of integral membrane proteins and a membrane peripheral network that includes only soluble proteins was observed (Fig. 8). The existence of two independent MICOS networks, composed of subunits whose stability depend on each other, suggests that the two subcomplexes are assembled independently. Regarding the functions of the two networks, it seems that the membrane-integrated subcomplex is mainly responsible for formation of CJs, whereas the most striking phenotype observed after the ablation of subunits of the peripheral subcomplex is an inhibition of mitochondrial protein import. Deletion or downregulation of individual subunits of the membrane-integrated MICOS network does not affect growth, indicating the formation of CJs is not essential for procyclic T. brucei 29-13, not even when it is grown in the absence of glucose which forces the cell to rely on oxidative phosphorylation for energy conversion (Lamour et al., 2005).
Ablation of individual subunits of the peripheral MICOS network in trypanosomes strongly interferes with mitochondrial protein import. Connections between protein import and MICOS have first been described in yeast, where it was shown that there are two different ways of how MICOS promotes protein import, both mediated by Mic60 (von der Malsburg et al., 2011; Bohnert et al., 2012; Wrobel et al., 2015). Mic60 facilitates membrane insertion of β-barrel proteins by its binding to the TOM and the SAM complexes, although the exact mechanism is unclear. Moreover, Mic60 transiently interacts with Mia40 and thus promotes import of IMS proteins by recruiting Mia40 to the vicinity of the TOM complex (von der Malsburg et al., 2011). Proteomics analyses of the TbMic34 RNAi cell line demonstrated that the earliest affected proteins are those localized in the IMS, including small Tim chaperones (Wenger et al., 2017), whose downregulation explains the general import phenotype that is observed at later time points. β-Barrel proteins, however, were not affected. Trypanosomes do not have a Mia40 orthologue (Basu et al., 2013) and their Mic60 orthologue is a member of the membrane integral subcomplex that is not essential for normal growth of the T. brucei 29-13 strain. The deficiency of IMS protein import that is seen after ablation of the essential membrane peripheral subcomplex can therefore not be connected to the lack of TbMic60. However, in T. brucei the function of Mic60 may be split between the Mic60 orthologue and TbMic34 (Kaurov et al., 2018), the core component of the peripheral network that may functionally compensate for the lacking mitofilin domain of TbMic60. In this regard the peripheral MICOS network may still contain a Mic60 remnant corresponding to the C-terminal half of the protein.
Nevertheless, we find it unlikely that TbMic34 is directly linked to the protein import phenotype. We prefer the previously proposed scenario, that the thioredoxin-like protein TbMic20 – a member of the peripheral MICOS subcomplex – might be a functional analogue of Mia40 and together with Erv1 directly mediate IMS protein import (Kaurov et al., 2018). Thus, protein import inhibition seen after the ablation of the peripheral MICOS network might ultimately be due to the concomitant depletion of TbMic20. However presently, indirect effects cannot be excluded and more evidence is required to show that TbMic20 indeed is a functional analogue of Mia40.
In summary, our study illustrates the value of a comparative analysis of the MICOS complex between opisthokonts and excavates. Together with future studies in other organisms this will provide us with an ‘evolutionary cell biology’ perspective (Brodsky et al., 2012; Lynch et al., 2014; Richardson et al., 2015) of the MICOS complex. This in turn should help us to determine which structural and functional features of the MICOS complex are universally conserved, not necessarily because of common descent but because of chemical and physical constraints imposed by functional selection.
Experimental procedures
Transgenic cell lines
All experiments were performed with procyclic T. brucei 29-13 and derivatives thereof (Wirtz et al., 1999). Double knockouts of TbMic10-1 and TbMic10-2 were generated by replacement of their ORFs of both alleles with the coding sequences of genes leading to resistance against phleomycin and puromycin. Growth of these cells was monitored in SDM-80 containing 10% (v/v) fetal calf serum supplemented with either 5.55 mM glucose (rich) or 50 mM N-acetylglucosamine (glucose-poor) (Lamour et al., 2005; Ebikeme et al., 2008). All other experiments were performed with cells cultured at 27°C in standard SDM-79 containing 10% FCS.
Tetracycline inducible RNAi against the ORFs of TbMic34 (nt 167–628), TbMic32 (nt 229–806), TbMic60 (nt 386–669) and TbMic20 (nt 54–504) was achieved by stable integration of NotI-linearized plasmids containing stem–loop constructs targeting the indicated sequences.
TbMic32 was tagged in situ with 3xHA (Oberholzer et al., 2005) in the background of RNAi against TbErv1 (Peikert et al., 2017). An additional cell line was made for inducible overexpression of a triple c-myc tagged TbMic32 from an ectopic copy encoded on a derivative of pLEW100 (Bochud-Allemann and Schneider, 2002).
Protein analysis
Digitonin extraction was used to separate cytoplasmic proteins from a crude mitochondrial pellet. In brief, 5 × 107 cells were harvested, washed in PBS and incubated in 20 mM Tris HCl pH 7.5, 0.6 M sorbitol, 2 mM EDTA and 0.015% (w/v) digitonin for 10 min on ice. Subsequently, the suspension was centrifuged for 5 min at 6,800 × g at 4°C and samples containing equal cell equivalents of whole cells, the supernatant and the mitochondria-enriched pellet were subjected to SDS-PAGE and immunoblot analysis.
For alkaline extraction, a mitochondria-enriched fraction was prepared as described above, resuspended in 100 mM Na2CO3 pH11.5 and incubated for 10 min on ice. Centrifugation at 100,000 × g for 10 min at 4°C separated the supernatant containing soluble proteins from pelleted integral membrane proteins.
For BN-PAGE analysis, a crude mitochondrial pellet was prepared by digitonin extraction (see above) from 1 × 107 cells. The pellet was then resupended in 100 µl of 20 mM Tris-HCl pH 7.4, 50 mM NaCl, 10% glycerol, 0.1 mM EDTA and 1.5% (w/v) digitonin and proteins were solubilized by incubation on ice for 20 min. After a clearing spin (15 min, 4°C, 21,000 × g) a fraction of the supernatant corresponding to 4 × 107 cells was separated on a 4-13 % BN-PAGE gradient gel. After electrophoresis the gel was incubated briefly in SDS-PAGE running buffer (25 mM Tris, 1 mM EDTA, 190 mM glycine, 0.05 % SDS) and proteins were blotted onto a PVDF membrane via semi-dry transfer.
Antibodies
Polyclonal antibodies against TbMic10-1 (used in a 1:1,000 dilution) and TbMic34 (diluted 1:2,500 for immunoblots) were obtained upon immunization of rabbits with a synthetic peptide (LRKGFRGSTGVSDRESSGS) or recombinant full-length protein expressed in E. coli respectively. All other antibodies are described in detail (Käser et al., 2017).
Transmission electron microscopy
Transmission electron microscopy on strain 29-13 and TbMic10-1 knockout cells was done as described (Schnarwiler et al., 2014) and pictures were analyzed with ImageJ. In short, uninduced and induced TAC40-RNAi cells of T. brucei were fixed in culture for at least 24 h, using 2.5% glutaraldehyde in 0.15 M Hepes buffer. Samples were postfixed with 1% (wt/vol) osmium tetroxide, dehydrated in ascending concentrations of ethanol, and embedded in Epon. Ultrathin sections were stained with uranyl acetate and lead citrate, using an ultrostainer (Leica), and observed in a Philips CM12 electron microscope at 80 kV equipped with a Morada camera system, using iTEM software.
Proteolytic digestion for MS analysis
Mitochondria-enriched fractions prepared from ΔTbMic10-1, ΔTbMic10-2 and control cells (110 µg of protein per sample) as well as from SILAC-labeled tetracycline-induced and uninduced TbMic34-RNAi cells (20 µg of protein per sample) were resuspended in 8 M urea/50 mM NH4HCO3. Cysteine residues were reduced using 5 mM Tris(2-carboxy-ethyl)phosphine/10 mM NH4HCO3 (incubation for 30 min at 37°C) and free thiol groups were subsequently alkylated with 50 mM iodoacetamide/10 mM NH4HCO3 (30 min at room temperature in the dark). Dithiothreitol was added to a final concentration of 20 mM to quench the alkylation reaction. Samples were diluted with 50 mM NH4HCO3 to a final urea concentration of 1.6 M and proteins were digested with trypsin (37°C, overnight). The reaction was stopped by adding acetic acid to a final concentration of 0.25% (v/v). Samples were desalted using StageTips and dried in vacuo (Rappsilber et al., 2007).
Peptide stable isotope dimethyl labeling and high-pH reversed-phase fractionation
Peptides derived from mitochondria-enriched fractions of ΔTbMic10-1, ΔTbMic10-2 and control cells were reconstituted in 100 mM tetraethylammonium bicarbonate (100 µl per 10 µg of protein). Peptides were labeled light, medium–heavy or heavy by stable isotope dimethyl labeling (Boersema et al., 2009) essentially as described previously (Peikert et al., 2017) including label switch. For light labeling, formaldehyde (CH2O) and sodium cyanoborohydride (NaBH3CN) were used, medium–heavy labeling was performed with deutered formaldehyde (CD2O) instead of the light variant, and for heavy labeling, 13C-containing deuterated formaldehyde (13CD2O) was used in combination with deuterated sodium cyanoborohydride (NaBD3CN). Labeling was performed by mixing 100 µl of reconstituted peptides with the respective isotopologue of formaldehyde (4% [v/v], 4 µl) and sodium cyanoborohydride (0.6 M, 4 µl) and incubation for 1 h at 20°C and 800 rpm. Reactions were stopped by addition of 16 µl of 1% NH3 and samples were subsequently acidified with 8 µl of 100% formic acid (FA). Differentially light, medium–heavy and heavy dimethyl-labeled peptides were mixed, purified using StageTips and dried in vacuo. Peptides were subsequently fractionated by high-pH reversed-phase chromatography on StageTips (Peikert et al., 2017). To this end, peptides were resuspended in 10 mM NH4OH and loaded onto StageTips equilibrated with 10 mM NH4OH. Peptides were fractionated by stepwise elution with 0%, 3%, 6%, 10%, 13%, 18%, 40% and 72% (v/v) ACN/10 mM NH4OH. Solvents and peptides were applied by centrifugation for 1–2 min at 800 × g. Peptides were dried in vacuo, reconstituted in 15 µl of 0.1% (v/v) TFA per fraction, of which 10 µl were analyzed by liquid chromatography–mass spectrometry (LC-MS).
LC-MS and analysis and data processing
Peptides corresponding to approximately 2 µg of protein were analyzed by nano-HPLC-ESI-MS/MS on an Orbitrap Elite (TbMic34-RNAi experiments) or a Q Exactive (ΔTbMic10-1/2 experiments) instrument (Thermo Fisher Scientific) each directly connected to an UltiMate 3000 RSLCnano HPLC system (Thermo Fisher Scientific). The RSLC system coupled to the Orbitrap Elite was equipped with PepMap C18 precolumns (5 mm × 300 µm inner diameter; Thermo Scientific) for preconcentration of peptides and C18 reversed-phase nano-LC columns (Acclaim PepMap RSLC columns; 50 cm × 75 µm inner diameter; particle size 2 µm, pore size 100 Å; Thermo Scientific) for peptide separation. The RSLC system coupled to the Q Exactive was operated with nanoEase M/Z Symmetry C18 precolumns (20 mm × 180 µm inner diameter; Waters) and a nanoEase M/Z HSS C18 T3 analytic column (25 cm × 75 µm inner diameter; particle size 1.8 µm, pore size 100 Å; Waters). Peptides of TbMic34-RNAi experiments were loaded onto the precolumns at a flow rate of 30 µl/min. The solvent system used for peptide separation consisted of 4% (v/v) DMSO/0.1% (v/v) FA (solvent A) and 48% (v/v) methanol/30% (v/v) ACN/4% (v/v) DMSO/0.1% (v/v) FA (solvent B). Peptides were eluted applying a gradient of 3%–60% solvent B in 315 min, 60%–95% B in 15 min at 5 min at 95%B at a flow rate of 250 nl/min and a column temperature of 40°C. Peptides of ΔTbMic10-1/2 experiments were loaded at a flow rate of 5 µl/min. Solvent system and gradient for peptide separation were as follows: 0.1% (v/v) FA (solvent A) and 86% (v/v) ACN (solvent B); 4%–40% B in 50 min, 40%–95% B in 5 min and 5 min at 95% B; flow rate and column temperature were 300 nl/min and 40°C respectively. MS data were acquired in data-dependent mode essentially as described before (Peikert et al., 2017).
Mass spectrometric raw data were processed using MaxQuant/Andromeda (version 1.5.5.1; (Cox and Mann, 2008; Cox et al., 2011)) and the fasta file for T. brucei TREU927 downloaded from the TriTryp database (version 8.1). MaxQuant default settings were used except that only one unique peptide and one ratio count were required for protein identification and relative quantification. For analysis of data from TbMic34-RNAi SILAC experiments, Lys0/Arg0 were set as light and Lys8/Arg10 as heavy labels; for data from peptide stable isotope dimethyl-labeling experiments (ΔTbMic10-1/2 experiments), dimethLys0/dimethNter0 was set as light, dimethLys4/dimethNter4 as medium–heavy and dimethLys8/dimethNter8 as heavy label. Data were visualized by plotting the mean of log10-tranformed normalized protein abundance ratios against the negative log10 of the corresponding p-value determined in a two-sided Student’s t-test. Information about proteins identified and quantified in ΔTbMic10-1/2 experiments and TbMic34-RNAi experiments are provided in Tables S1 and S2A and B, respectively.
Miscellaneous
Knockdown efficiency of RNAi against TbMic60 was analyzed by RT-PCR. Briefly, total RNA was isolated and reverse transcribed using oligo-dT primers. Resulting cDNA was amplified using a TbMic60-specific reverse primer and a forward primer hybridizing to the spliced leader sequence ensuring that PCR products can only derive from cDNA and not from traces of genomic DNA. In vitro import assays were done as described (Hauser et al., 1996), using in vitro translated CoxIV precursor as a substrate.
Acknowledgements
This study was supported by the NCCR ‘RNA & Disease’ (to AS) and in part by grants 175563 (to AS) all funded by the Swiss National Science Foundation. Work in the lab of BW was supported by the German Research Foundation (DFG) under Germany’s Excellence Strategy (CIBSS – EXC-2189 – Project ID 390939984). Support from the Czech Science Foundation grant 17-24036S (to HH) as well as ERC CZ LL1601 and the ERD Funds, project OPVVV 16_019/0000759 (to JL) is acknowledged. Electron microscopy sample preparation and imaging were performed with devices supported by the Microscopy Imaging Center (MIC) of the University of Bern.
Author contributions
CE, JB, JM, BS designed and performed experiments and analyzed data. SO and BW produced and analyzed the proteomic MS results. BH and BZ performed the EM analysis. JL and HH conceived experiments. All authors edited the paper. AS wrote the paper, supervised the work and obtained the main funding.
Conflict of interest
The authors declare that they have no conflict of interest.