Geographical contrasts of Y-chromosomal haplogroups from wild and domestic goats reveal ancient migrations and recent introgressions
Abstract
By their paternal transmission, Y-chromosomal haplotypes are sensitive markers of population history and male-mediated introgression. Previous studies identified biallelic single-nucleotide variants in the SRY, ZFY and DDX3Y genes, which in domestic goats identified four major Y-chromosomal haplotypes, Y1A, Y1B, Y2A and Y2B, with a marked geographical partitioning. Here, we extracted goat Y-chromosomal variants from whole-genome sequences of 386 domestic goats (75 breeds) and seven wild goat species, which were generated by the VarGoats goat genome project. Phylogenetic analyses indicated domestic haplogroups corresponding to Y1B, Y2A and Y2B, respectively, whereas Y1A is split into Y1AA and Y1AB. All five haplogroups were detected in 26 ancient DNA samples from southeast Europe or Asia. Haplotypes from present-day bezoars are not shared with domestic goats and are attached to deep nodes of the trees and networks. Haplogroup distributions for 186 domestic breeds indicate ancient paternal population bottlenecks and expansions during migrations into northern Europe, eastern and southern Asia, and Africa south of the Sahara. In addition, sharing of haplogroups indicates male-mediated introgressions, most notably an early gene flow from Asian goats into Madagascar and the crossbreeding that in the 19th century resulted in the popular Boer and Anglo-Nubian breeds. More recent introgressions are those from European goats into the native Korean goat population and from Boer goat into Uganda, Kenya, Tanzania, Malawi and Zimbabwe. This study illustrates the power of the Y-chromosomal variants for reconstructing the history of domestic species with a wide geographical range.
1 INTRODUCTION
Goat (Capra hircus), sheep (Ovis aries), cattle (Bos taurus) and pigs (Sus scrofa) are four major livestock species, which after their domestication in southwest Asia about 10,000 years ago (Larson & Fuller, 2014; Stiner et al., 2022) spread to all inhabited continents. Because of their relatively small size, sheep and goats were the earliest domesticates, but have become less important than cattle and pigs as suppliers of food. However, sheep and goat are suitable for extensive management by smallholders or hobby breeders with goats being favoured in conditions of poverty (Peacock, 2005). Although the high-quality goat cashmere wool and mohair fibres do not attain the volume of sheep wool, the demand for goat milk and cheese has increased considerably since the 1960s (Dubeuf et al., 2004; Miller & Lu, 2019). In the last four decades, this has doubled the global number of goats to around one billion (Utaaker et al., 2021), approaching the numbers for sheep and cattle (Hegde, 2019).
As for other livestock, genetic isolation, adaptation and selection have created numerous local goat populations, whereas a restricted number of high-performing breeds play a major role in agricultural production. The genetic diversity of goat breeds has been studied extensively (Ajmone-Marsan et al., 2014; Amills et al., 2017; Deniskova et al., 2021; Zheng et al., 2020). This demonstrated for autosomal DNA a geographical partitioning of the diversity (Colli et al., 2018), which is in sharp contrast to similar studies of sheep (Belabdi et al., 2019; Ciani et al., 2020; Kijas et al., 2012), but has been confirmed based on similarities of ancient and modern DNA samples from the same regions (Cai et al., 2020; Daly et al., 2018, 2021). However, the goat Y-chromosome as a marker for paternal lineages has not yet been studied at a worldwide scale.
Because of an absence of recombination, the male-specific part of the mammalian Y-chromosome is by far the longest haplotype that is stably transmitted across generations (Hughes et al., 2015). In many species, males have a relatively small male effective population size, which makes Y-chromosomal variants highly informative markers for paternal origin that generally show a much stronger phylogeographical differentiation than mitochondrial or autosomal variants. This is now widely exploited in population-genetic studies of humans (Batini & Jobling, 2017; Jobling & Tyler-Smith, 2017; Kivisild, 2017), cattle (Edwards et al., 2011; Ganguly et al., 2020; Xia et al., 2019), horse (Felkel et al., 2019a; Wallner et al., 2017; Wutke et al., 2018), water buffalo (Zhang et al., 2016), sheep (Deng et al., 2020; Meadows & Kijas, 2009; Zhang et al., 2014), camel (Felkel et al., 2019b), llamas and alpacas (Marín et al., 2017), pigs (Choi et al., 2020; Guirao-Rico et al., 2018) and dogs (Natanaelsson et al., 2006; Oetjens et al., 2018).
A preliminary analysis of the Y-chromosomal diversity in European and Turkish goats defined the three haplotypes, Y1A, Y1B and Y2, which had a strong geographical differentiation (Lenstra & Econogene Consortium, 2005). The same haplotypes were found in goats from Portugal and North Africa (Pereira et al., 2009), Turkey (Çinar Kul et al., 2015), eastern and southern Asia (Tabata et al., 2018, 2019; Waki et al., 2015), and Switzerland and Spain (Vidal et al., 2017), with the additional haplotypes Y2B in east Asia, Y2C in Turkish Hair and Kilis goats, and Y1B2 as well as Y1C mainly in Switzerland (Table S1). However, these haplotypes are based on a low number of single nucleotide polymorphisms (SNPs) in or near DDX3Y, SRY and ZFY and genotyping in a limited number of domestic goat breeds. Thus, it is not clear if the haplotypes represent major haplogroups or local variants or if other major haplogroups exist. Nor does it inform us on the Y-chromosomal variants that existed in earlier domestic goats or in their wild ancestor, the bezoar (Capra aegagrus; Amills et al., 2017). Whole-genome sequencing (WGS), however, has confirmed the differentiation of the Y1 and Y2 haplogroups (Xiao et al., 2021; Zheng et al., 2020).
In this study, we used WGS data for a large panel of goat breeds (Denoyelle et al., 2021) to systematically characterize the SNP-level variation in the single-copy male-specific part of the caprine Y-chromosome. In addition, we determined the Y-chromosomal haplogroups in goats originating from several European, Asian or African countries, in ancient goat DNA samples and in the wild bezoar (Alberto et al., 2018; Cai et al., 2020; Daly et al., 2018, 2021; Zhang et al., 2014; Zheng et al., 2020). We sought to answer the following questions: (i) How are the WGS-based haplogroups related to the previously reported haplotypes? (ii) How are the domestic paternal lineages related to those of bezoars from Iran and Anatolia, respectively? (iii) How strong is the phylogeographical structure of the caprine male lineages? (iv) What does the pattern of diversity tell us about Neolithic and later migrations? (v) Can we also infer other gene flows between or within continents? Answering these questions will contribute to our understanding of the genetic background of the domestic goat, which is relevant for breed management and conservation.
2 MATERIALS AND METHODS
2.1 WGS data, filtering and phylogenetic analysis
We selected as source of the SNPs four scaffolds that together cover 1,567,760 bp of the male-specific part of the caprine Y-chromosome. These are unplaced in the ARS1 assembly but for a large part closely match a recent Y-chromosomal contig of the Saanen_v1 assembly (Table S2; Li et al., 2021) and contain the single-copy Y-chromosomal genes SRY, DDX3Y and ZFY and the SNPs that define the major haplotypes Y1A, Y1B, Y2A and Y2B (Çinar Kul et al., 2015; Lenstra & Econogene Consortium, 2005; Waki et al., 2015; see Table S1). The genes USP9Y, UTY, DDX3Y and ZFY are proximate near one of the ends of the male-specific Y-chromosomal region and well separated from SRY (Li et al., 2021). The selected contigs have a low overall level of apparent heterozygosity, indicating a high frequency of hemizygous markers (Table S2).
In a preliminary study (https://www.biorxiv.org/content/biorxiv/early/2020/02/17/2020.02.17.952051.full.pdf), we used WGS data from the Sequence Read Archive (SRA) for 70 mainly Asian and Moroccan male goats (Alberto et al., 2018; Zheng et al., 2020; Table S3). We extracted the genotypes of 5356 SNPs as described (Zheng et al., 2020), which after filtering yielded 2350 SNPs without female- of male-heterozygous scores, <5% missing scores/SNP and a minor allele frequency (MAF) >0.02.
- After removal of indels, 54,032 Y-chromosomal SNPs were retained.
- From the 948 female goats, 670 were selected with scores of <1% for the 54,032 Y-chromosomal SNPs in order to minimize scores due to contamination with male DNA.
- In total, 17,228 SNPs were scored in at least one of the 670 females and were removed.
- From the 36,804 remaining male-specific SNPs, 7263 SNPs had ≥1 heterozygous score and 506 were monomorphic in 424 male goats, keeping 29,035 hemizygous SNPs.
- From the 380 domestic male samples, 354 with a call rate of >95% were kept. From the 34 wild goats, two Italian ibexes and four Iranian bezoars had call rates of only 90.7%–94.4%, but this did not appear to affect their phylogenetic positions or the corresponding bootstrapping values (see below), so these were retained in the data set.
- From the 29,035 SNPs, 12,540 had a call rate of <99% in the 388 male goats (354 domestic and 34 wild) and were discarded.
- From the remaining 16,495 SNPs, 552 SNPs had an MAF in the 354 male domestic goats of >1% and from the other 15,943, 9977 SNPs had a at least one score in wild goats, totalling 10,529 SNPs representing male-specific Y-chromosomal variation in domestic and/or wild goats, which differentiate 27 wild and 80 domestic haplotypes.
- Finally, two domestic goats with unknown breed origins were discarded, resulting in a final panel of 386 (352 domestic and 34 wild) goats.
Allele-sharing distances between individuals were calculated using plink or mega7 (Tamura et al., 2011), and visualized in neighbour-joining (NJ) trees by using the program splitstree4 (Huson & Bryant, 2006). For calculating bootstrapping values in an NJ tree of the 80 domestic and 20 bezoar haplotypes and as an outgroup one markhor, we selected 2867 SNPs with MAF >5% and used the program mega7. The topology of this tree was essentially identical to a tree of genotypes of the 10,529 SNP panel for the same samples.
For construction of median-joining networks (Bandelt et al., 1999), we selected 286 male goats and 27 bezoars, omitting other wild goats as well as transboundary breeds outside their region of origin and balancing the breed representation by analysing ≤18 individuals per breed. With the number of polymorphic SNPs thus reduced to 1734, the program popart (Leigh & Bryant, 2015; http://popart.otago.ac.nz) generated a network of 91 haplotypes (Table S4).
In addition to the SNPs previously by dideoxy sequencing (Çinar Kul et al., 2015; Lenstra & Econogene Consortium, 2005; Vidal et al., 2017; Waki et al., 2015), we identified other diagnostic SNPs in the VarGoats data set on the basis of an FST genetic distance (plink version 1.9 –fst) of 1.0 between males with a given haplogroup and all other males. Several Y-chromosomal diagnostic SNPs have been incorporated in the Goat_IGGC_65K_v2 bead array (Table S1).
2.2 Haplogroups distribution in 186 goat breeds
- From the goat panel collected for the Econogene project (Lenstra & Econogene Consortium, 2005), DNA samples of 353 male goats from 38 European or southwestern Asian breeds were analysed by PCR (polymerase chain reaction) amplification and dideoxy-sequencing of DDX3Y, SRY and ZFY segments as described previously for bovine samples (Edwards et al., 2011; Nijman et al., 2008) using goat-specific primers (Table S6).
- We used published data from five Portuguese breeds or from Moroccan goats (Pereira et al., 2009), 12 Asian breeds or national populations (Waki et al., 2015, Tabata et al., 2018, 2019, combined with unpublished data), eight Turkish breeds (Çinar Kul et al., 2015), and 26 Spanish and Swiss breeds (Vidal et al., 2017).
- Haplogroup assignments for 354 domestic male goats in the VarGoats data set were derived from their positions in the phylogenetic trees (Figure 1b; Figure S2), which were fully consistent with the alleles of the diagnostic SNPs (Table S1). This implies that they represent the variation that corresponds to the basal branch of the respective haplogroups in the phylogenetic trees or networks (Figures 1b and 2). Haplogroups for VarGoats male goat with a call rate of <95% were inferred from the alleles of diagnostic SNPs (Table S1).
- In total, 368 DNA samples from several sources, including the AdaptMap panel (Colli et al., 2018) were genotyped by the KASP assay (Kompetitive allele specific PCR assay) for SNPs NW_017189563.1 g.T280306>A and NW_017189885.1 g.A11686>G, carried out at the Van Haeringen Laboratory (Wageningen, Netherlands).These SNPs differentiate Y1 vs. Y2 and Y1A vs. Y1B, respectively.
- For 31 breeds, genotypes of diagnostic SNPs (Table S1) were obtained by megablast searching of SRA entries with queries of 40–50 bp overlapping the SNPs. SRA data for pools of individuals were only used if this allowed an unambiguous identification of the haplogroup composition.
For samples collected by Vidal et al. (2017) and Çinar Kul et al. (2015) and for samples analysed by KASP before genomic data became available, Y1AA and Y1AB have been both scored as the Y1A haplotype. Several of these samples did not contain Y1A or belonged to breeds for which additional data are available (Table S5). However, for 17 breeds we only have the Y1A (Y1AA + Y1AB) frequency. Likewise, Vidal et al. (2017) and the KASP assays did not differentiate Y2A and Y2B. Since the VarGoats and Econogene panels with comprehensive coverage of Europe and Africa did not contain a single Y2B-carrying goat, we assigned Y2 scores in other European and African goats to Y2A.
2.3 Haplotypes of ancient DNA samples
Daly et al. (2021) assigned ancient DNA (aDNA) samples from southwest Asia and southeast Europe to the domestic Y-chromosomal haplogroups on the basis of their positions in a phylogenetic tree. Cai et al. (2020) and Zheng et al. (2020) described eight Chinese aDNA samples and one medieval sample from the northern Caucasian region. Sample YJL2G (coverage 13.4×) clustered with the Y1AB haplogroup, but the coverage of the others ranged from 0.013 to 0.118×. This resulted in insufficient overlap with the 10,529 male-specific hemizygous SNPs in domestic goats (see above). Therefore, we relaxed our filtering and allowed SNPs scoring in up to 1% of the females. We excluded SNPs with male heterozygote scores only if these occurred in a panel of individuals with call rates >95% representing the 80 different domestic haplotypes (see above) but removing two individuals with a high heterozygosity. This resulted in 5593 SNPs, 1018 of which were also scored in the low-coverage aDNA samples. The combined phylogenetic signals (Table S7) showed for sample GTM6G a high proportion of inconsistent scores, presumably due to contamination, but allowed plausible haplogroup assignment for samples KA1G, SMG1, SMG7, SMG11 and YJL2G and a tentative assignment for BG3 (Table S7).
3 RESULTS
3.1 Phylogeny of Y-chromosomal haplogroups
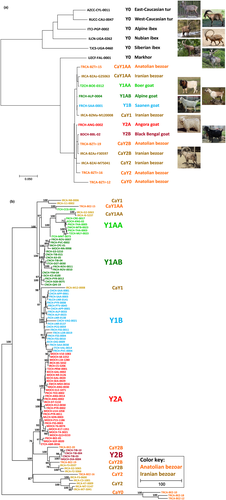
A phylogenetic tree of the wild and domestic goats (Figure 1a) shows an intermingling of bezoars and the domestic goat. From the other wild goat species, the markhor is the closest relative of the bezoar and the domestic goat.
We found 107 different haplotypes in our panel of 352 domestic goats and 27 bezoars with markhor as an outgroup. A phylogenetic tree (Figure 1b) shows haplogroups that correspond to the haplotypes Y1B, Y2A (Lenstra & Econogene Consortium, 2005) and Y2B (Waki et al., 2015) whereas the Y1A haplotypes are split into haplogroups Y1AA and Y1AB. This is confirmed in a data set of mainly Asian and Moroccan goats (Figure S1). All domestic haplotypes differ from the Iranian or Anatolian bezoar haplotypes, which also differ from each other. The bezoar haplotypes are associated with the domestic Y1AA or Y2B clusters (CaY1AA and CaY2B, respectively), are linked to the Y1 or Y2 roots (CaY1 and CaY2) or are outside the domestic cluster (Y0).
Figure S2 shows subtrees containing all 352 domestic goats. This figure also indicates goats with the previously proposed local haplotypes (Vidal et al., 2017; this study, Table S1): Y1AB2 (this study), Y1B2 and Y1C. Y1B2 is represented by Swiss, French and Dutch goats. Our goat panel does not contain goats with the Y2C diagnostic allele (Çinar Kul et al., 2015).
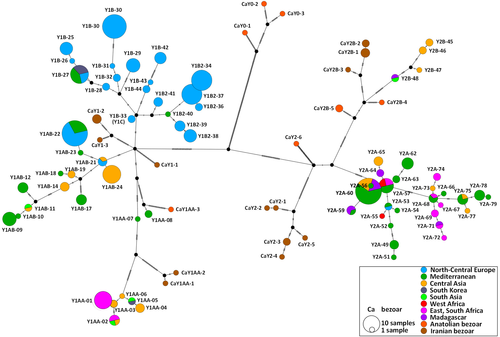
The phylogenetic relationships of domestic and bezoar haplotypes are confirmed by a median-joining network (MJN, Figure 2). Both the NJ tree and the MJN allow a few interesting observations to be made: (i) a close relationship of Y1B sequences from Switzerland and Korean Native goats, suggesting recent crossbreeding; (ii) likewise, a close relationship of Y1AA sequences from Central as well as South Asia, from South African Boer goats and from other Y1AA goats in southern and eastern Africa; and (iii) a clear divergence of the Malagasy Y2A haplotypes from the African continental haplotypes.
3.2 Geographical distribution of haplogroups
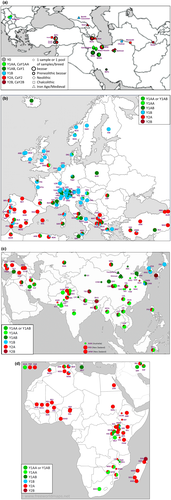
- Haplogroup Y1B is predominant in central and northern Europe, but outside Europe and North Africa it has only been found in one Ugandan Karamonja goat, in the Korean native breed and in exported Saanen populations.
- In northern and central Europe Y2A is only found in a single French des Fossés sample and together with Y1AA in the crossbred Anglo-Nubian. It is the predominant haplogroup in Spain, Anatolia and Africa south of the Sahara, but it is not found in China or Southeast Asia except in about 20% of the goats in the Philippines and Sulawesi. Remarkably, most Y2A haplotypes on Madagascar are more closely related to Asian than to continental-African Y2A haplotypes.
- Y2B is absent in Europe, continental Africa and west Asia, but is a major haplotype in east and southeast Asia. It is also observed in one Malagasy Diana sheep.
- Y1AA in Europe is only represented by three haplotypes in the local Ciocara breed and the Montecristo island population (Somenzi et al., 2022). The Italian haplotypes are outside the cluster of closely related south Asian and southeast African Y1AA haplotypes (Figure 2; Figure S2).
- The available data suggest a contrast of Y1AB dominating in northern China and Y1AA with Y2B in the south.
4 DISCUSSION
One of the benefits of the current availability of WGS data sets is the access to an abundance of sequence variants, which allow a comparison of individuals or populations for several purposes. This is especially useful for the analysis of Y-chromosomal diversity, which was previously restricted by the availability of Y-chromosomal markers. The male-specific part of the Y-chromosome constitutes the longest haplotype in the mammalian genome and may serve as a marker for mammalian paternal lineages. Here, we combined the data set of the VarGoats project with published data and genotyped diagnostic SNPs in male goat samples from several sources.
The Y-chromosomal phylogeny of wild and domestic goats is in agreement with the Y-chromosomal tree on the basis of AMELY and ZFY gene fragments (Pidancier et al., 2006) and with a phylogeny of WGS sequences (Grossen et al., 2020; Cai et al., unpublished). Mitochondrial DNA (mtDNA) trees confirm the close relationship of markhor (Capra falconeri) with bezoar and domestic goat, but do not show the separation of these species and the other wild goats. In addition, mtDNA sequences of some, but not all East Caucasian turs (Capra cylindricornis) cluster with the mtDNA sequences of markhor, bezoar and domestic goat, illustrating a separate history of maternal and paternal lineages in cross-fertile species (Chen et al., 2018; Marín et al., 2017; O'Connell et al., 2014; Zhang et al., 2016, 2020).
On the basis of WGS data, Zheng et al. (2020) and Xiao et al. (2021) reproduced the divergence of the domestic Y1 and Y2 haplogroups previously found on the basis of SNPs within or near Y-chromosomal genes (Lenstra & Econogene Consortium, 2005). Here we report a further differentiation of haplogroups, resulting in a phylogeny supported by a largely independent preliminary data set (Figure S1) and two phylogenetic algorithms. We found that the major haplogroups correspond to haplotypes defined by SNPs (Çinar Kul et al., 2015; Lenstra & Econogene Consortium, 2005; Pereira et al., 2009; Tabata et al., 2018, 2019; Vidal et al., 2017; Waki et al., 2015), but Y1A haplotypes belong to either haplogroup Y1AA or Y1AB.
The phylogeny also indicates that these haplogroups diverged after the split of the markhor and the cluster of the wild bezoar and domestic goats. The domestic goats and the two bezoar populations from Anatolia and Iran do not share haplotypes, whereas the bezoar haplotypes are attached to deep nodes in the tree of mainly domestic haplotypes. This suggests an absence of male gene flow between the bezoar populations and between the bezoar and domestic goats from the same region. Thus, domestic goats, which possibly were derived from bezoar populations not sampled in this study, maintained their paternal lineages during migration from the Fertile Crescent via Anatolia to Europe, despite indications of management of wild goats in central Anatolia (Stiner et al., 2022).
- The dominance of haplogroup Y1B in central and northern Europe may very well reflect population bottlenecks during the Neolithic introduction of agriculture via the Danube route (Cymbron et al., 2005; Rivollat et al., 2015; Tresset & Vigne, 2007)
- Y2A and Y1AA are almost the only haplogroups in Africa south of the Sahara. The two African Y1AA haplotypes are related to those of Asia, indicating that only Y2A expanded during the first introduction of domesticated goats in central and southern Africa
- Y2B has been found in two Neolithic Iranian samples whereas related CaYB2 haplotypes are present in Iranian and Anatolian bezoars. However, as a result of population bottlenecks during the global spread of domestic goats, Y2B now occurs only in Asia east of the Indus River and in one goat from Madagascar (see below).
- Y1AA was found in Neolithic samples in southeast Europe, but now has a low frequency in Europe. In Asia it expanded together with Y2B and later came to South Africa when Asian goats were used to breed the Boer goat (see below).
- Y1AB is the most frequent haplogroup in north China. The distributions of Y1AB and Y1AA/Y2B in East Asia correspond to ranges of the north Chinese cashmere goats and the small Southeast Asian “katjang” type, respectively (Porter et al., 2016). This obviously reflects the large difference in climate between northern and southern China, which determined a similar distribution of taurine and indicine cattle. These two types of cattle are considered to have entered China via a northern and southern migration route, respectively (Chen et al., 2018; Zhang et al., 2020), supporting the separate eastern expansions of the Y1AB and Y1AA/Y2B goats, respectively.
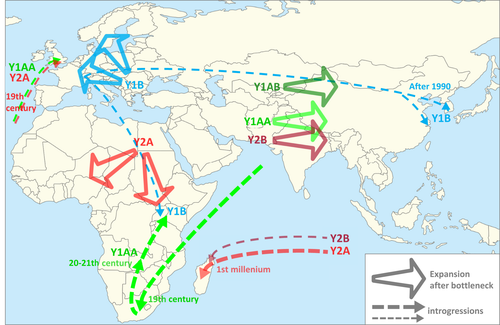
Exceptions to these geographical patterns follow from close relationships between haplotypes from different continents, which are probably explained by later major introgressions. Interestingly, in the phylogenetic trees and networks (Figure 2; Figure S2) the Y2A haplotypes on Madagascar are closely related to Asian haplotypes and one Diana goat from northern Madagascar even has an eastern Asian Y2B haplotype. However, autosomal DNA shows that the Malagasy goats are more closely related to the southern and eastern African continental goats (Colli et al., 2018; Denoyelle et al., 2021). This parallels a recent finding that Malagasy cattle combine Indian and admixed African zebu ancestry (Magnier et al., 2022). The Malagasy language has an Austronesian origin, which testifies to the colonization of Madagascar by immigrants from southeastern Asia about 500 CE. Thus, it is likely that these immigrants brought Austronesian goats, cattle and possibly also other livestock from their region of origin to Madagascar.
Other introgressions are more recent. The exceptional Y1AA and Y2A in the English Anglo-Nubian is explained by the documented import during the 19th century of Indian and African goats to England. These served on the ships as sources of milk and meat, but surviving males were crossed with English goats, which resulted in the emergence of a popular transboundary breed.
The worldwide popular Boer goat also is of mixed origin (Porter et al., 2016; Vidal et al., 2017) and carries exclusively Y1AA haplotypes. This breed is considered to be a crossbred of local African and Indian goats, possibly mediated by incrossing of Anglo-Nubian males (Porter et al., 2016). The crossbred origin is consistent with the results of Colli et al. (2018): a separate phylogenetic position of the Boer relative the other African and Asian goats and a K = 3 pattern of model-based clustering showing African and west Asian ancestry. The Indian ancestry is entirely in agreement with a close clustering of the Boer and Pakistani Y1AA haplotypes (Figures 1b and 2).
Subsequently, the Boer became itself a source of introgression. The same Y1AA haplotypes are closely related to Y1AA haplotypes in local breeds in Uganda, Malawi, Mozambique and Zimbabwe. In these countries crossbreeding with Boer goats from Africa is popular because of its excellent meat production (Banda et al., 1993; Garrine, 2007; Lu, 2011; Onzima et al., 2018). Therefore, it is most likely that the Y1AA haplotype in eastern and southern African goats originates from the Boer goat.
There were three out-of-range findings of Y1B, in the Ugandan Karamonja, in the Korean native goat and in the indigenous goats kept on Chongmin Island in Shanghai. Because of the popularity of Swiss dairy goats in both Uganda (NAADS, 2005) and Korea (Kim et al., 2019), crossbreeding again is the most likely explanation. Although European admixture in the Chongmin goats (Gao et al., 2020) has not been reported, the exotic occurrence of Y chromosomal variants appears to be a direct and sensitive indicator of admixture events. These need to be complemented with quantitative admixture tests, such as model-based clustering, the f3 and f4 test or, ideally, identification of introgressed segments across the genome.
The latter approach may also lead to clues regarding the phenotypic consequences of introgression via the identification of the admixed genes (Chen et al., 2018; Lv et al., 2014; Wang et al., 2015; Zheng et al., 2020). A more direct link with Y-chromosomal variation would be provided if this can be linked to male phenotypic traits, but even in human genetics this has scarcely be investigated (Matsunaga et al., 2021; Yang et al., 2018; Zhang et al., 2021). Breeds in which different Y-chromosomal haplogroups occur may allow us to study an association of haplogroups with typically male traits such as male fertility and dominance behaviour. It would be interesting to see if Y-chromosomal variants can be related to climate or other environmental features, because this would imply that the geographical differentiation of the Y-chromosomal variation is driven by regional adaptation.
Most introgressions described in this study contribute to the expansion of popular breeds at the expense of the original local breeds. On the one hand, depending on the extent of gene flow this may decrease the diversity of the genetic resources; on the other hand, it does not necessarily disrupt the environmental adaptation, arguably one of the most important components of the phenotypic repertoire. If properly managed, admixture of productive breeds may also contribute to the sustainable conservation of local populations and illustrates that genetic diversity has never been a static phenomenon.
We conclude that the Y-chromosomal variation of goats reveals bottlenecks, expansions and introgressions, illustrating the power of Y-chromosomal markers for inferring the genetic origin of mammalian populations.
AUTHOR CONTRIBUTIONS
IJN, PAM and JAL designed the study; GT-K and LC coordinated the VarGoats project; IJN and BÇK carried out the ABI sequencing; BDR, PB, TF, ZZ, YJ, YC, ZC, MMi and GS analysed the WGS data; TC, FPo and CD supplied the bezoar genotypes; KGD, DGB, YC and YJ supplied the aDNA data; HM, FK, SS, MMa, YN, AA, JSM, IAD, SRAB, FMD, TD, MKS, MB and PK provided genotypes for south Asian goats; BDR supplied most of the African samples for KASP genotyping; VAB, DB, BB, TB, SC, VC-C, LD, JG, JH, JK, NKh, NK, AM, RM, JMc, NAOC, FPe, AdS, MS, JS, AS, JT and HZ collected material and/or data for other breeds; JAL and IJN performed the downstream analysis; JAL wrote the first draft; and KGD, AM, FPe, BÇK, JMc, MM, MS, PAM, LC, CD and GTK contributed to the text.
ACKNOWLEDGEMENTS
The VarGoats project sequencing effort has been funded mainly by France Génomique “Call for high impact projects” (ANR-10-INBS-09-08) and by the BBSRC Global Challenges Research Fund Data and Resources Grant BBS/OS/GC/000012F awarded to the Roslin Institute for Reference genome and population sequencing of African goats. Detailed funding sources of the VarGoats project are described in Denoyelle et al. (2021). This study was supported by the Croatian Science Foundation (Project ANAGRAMS-IP-2018-01-8708) “Application of NGS in assessment of genomic variability in ruminants” and by the European Union (projects ECONOGENE QLK5–CT2001–02461). We are grateful to Dr E. Cuppen (Utrecht Medical Centre) for access to dideoxy sequencing facilities.
CONFLICT OF INTEREST
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
VarGoats CONSORTIUM
Australia: James Kijas, CSIRO. Denmark: Bernt Guldbrandtsen, Aarhus University; Finland: Juha Kantanen, Luke. France: Gwenola Tosser-Klopp, Philippe Bardou, Laure Denoyelle, Thomas Faraut, Julien Sarry, Estel le Talouarn, GenPhySE, Castanet-Tolosan; Adriana Alberti, Céline Orvain, Stefan Engelen, Université Paris-Saclay, Evry; Dylan Duby, Museum National d'Histoire Naturelle; Pierre Martin, Capgenes; Coralie Danchin, Delphine Duclos, Institut de l'Elevage; Daniel Allain, Remy Arquet, Nathalie Mandonnet, Michel Naves, Isabelle Palhiere, Rachel Rupp, INRAE, and CABRICOOP breeders; Francois Pompanon, LECA. Iran: Hamid R. Rezaei, Gorgan University of Agricultural Sciences and Natural Resources. Ireland: Sean Carolan and Maeve Foran, Old Irish Goats Society. Italy: Alessandra Stella, IBBA-CNR; Paolo Ajmone-Marsan, Licia Colli, Marcello Del Corvo, Universita Cattolica del Sacro Cuore; Alessandra Crisa, Council for Agricultural Research and Economics (CREA); Donata Marletta, University of Catania, Italian Goat Consortium; Paola Crepaldi, University of Milan, Italian Goat Consortium; Michele Ottino, Parco Nazionale del Gran Paradiso; Ettore Randi, ISPRA Istituto Superiore per la Protezione e la Ricerca Ambientale. Kenya: Denis Fidalis Mujibi, International Livestock Research Institute (ILRI), Nairobi; Malawi: Timothy Gondwe, Department of Animal Science, Lilongwe University of Agriculture and Natural Resources. Morocco: Badr Benjelloun, INRA Maroc. Mozambique: Maria Da Gloria Taela, Animal Production Institute, Ministry of Agriculture, Maputo. Netherlands: Johannes A, Lenstra, Utrecht University. Nigeria: Oyekan Nash, National Biotechnology Development Agency. Pakistan: Muhammad Moaeen-ud-Din, PMAS-Arid Agriculture University. South Africa: Carina Visser, Faculty of Natural and Agricultural Sciences. Spain: Felix Goyache, Isabel Alvarez, Área de Genética y Reproducción Animal del Serida, Villaviciosa; Marcel Amills and Armand Sànchez, Centre for Research in Agricultural Genomics (CRAG); Juan Capote, Instituto Canario de Investigaciones Agrarias (ICIA); Jordi Jordana, Universitat Autònoma de Barcelona; Agueda Pons, Serveis de Millora Agrària i Pesquera (SEMILLA), Illes Balears; Amparo Martínez and Antonio Molina, University of Córdoba. Switzerland: Cord Drögemüller, University of Bern. Tanzania: Hassan Ally Mruttu, Ministry of Livestock and Fisheries Development. Uganda: Clet Wandui Masiga, Tropical Institute of Development Innovations (TRIDI); UK: Emily Clark, Mazdak Salavati, University of Edinburgh, Edinburgh. USA: Benjamin Rosen, Curtis P. Van Tassell, USDA/ARS; Jim Reecy, Iowa State University, Ames; Gordon Luikart, Montana Conservation Genomics lab (MCGL), Division of Biological Sciences, University of Montana, Missoula. Zimbabwe: Joseph Sikosana, Department of Research and Specialist Services, Division of Livestock Research.
ECONOGENE CONSORTIUM
Albania: Hoda Anila, Dobi Petrit, Fac. Agriculture, Tirana. Austria: Baumung Roswitha, Univ. Natural Resources Applied Life Sciences, Vienna. Belgium: Baret Philippe, Fadlaoui Aziz, Univ. Cath. Louvain, Louvain-la-Neuve. Cyprus: Papachristoforou Christos, Agricult. Research Instit., Nicosia. Egypt: El-Barody M.A.A., Minia Univ. France: Taberlet Pierre, England Phillip, Luikart Gordon, Beja-Pereira Albano, Zundel Stéphanie, Univ. Joseph Fourier, Grenoble; Trommetter Michel, Inst. Recherche Agronomique, Grenoble. Germany: Erhardt Georg, Brandt Horst, Ibeagha-Awemu Eveline, Lühken, Gesine, Daniela Krugmann, Prinzenberg Eva-Maria, Lipsky Shirin, Gutscher Katja, Peter Christina, Justus-Liebig Univ. Giessen; Roosen Jutta, Bertaglia Marco, Univ. Kiel. Greece: Georgoudis Andreas, Al Tarrayrah, Jamil, Kliambas Georgios, Kutita Olga, Karetsou Katerina, Aristotle Univ. Thessaloniki, Thessaloniki; Ligda Christina, National Agric. Research Foundation, Thessaloniki. Hungary: Istvan Anton, Fesus Lazlo, Research Inst. Animal Breeding Nutrition, Herceghalom. Italy: Ajmone-Marsan Paolo, Canali Gabriele, Milanesi Elisabetta, Pellecchia Marco, Università Cattolica S. Cuore, Piacenza; Carta Antonello, Sechi Tiziana, Ist. Zootecnico Caseario Sardegna, Sassari; Cicogna Mario, Fornarelli Francesca, Giovenzana Stefano, Marilli Marta, Univ. Studi di Milano; Marletta Donata, Bordonaro S., D'Urso Giuseppe, Univ. Studi Catania; Pilla Fabio, D'Andrea Mariasilvia, Univ. Molise, Campobasso; Valentini Alessio, Cappuccio Irene, Pariset Lorraine, Univ. Tuscia, Viterbo. Jordan: Abo-Shehada Mahamoud, Jordan Univ. Science Technology. Netherlands: Lenstra Johannes A., Nijman, Isaäc J., Van Cann, Lisette M., Utrecht Univ. Poland: Niznikowski Roman, Dominik Popielarczyk, Strzelec Ewa, Warsaw Agricultural Univ. Romania: Vlaic Augustin, Univ. Cluj-Napoca. Spain: Dunner Susana, Canon Javier, Cortes Oscar, Garcia David, Univ. Complutense Madrid. Switzerland: Caloz Régis, EPFL, Lausanne; Obexer-Ruff Gabriela, Marie-Louise Glowatzki, Universität Bern. Turkey: Ertugrul Okan, Ankara Univ.; Togan Inci, Koban Evren, Middle East Technical Univ., Ankara. UK: Bruford Mike, Perez Trinidad, Juma Gabriela, Cardiff Univ.; Hewitt Godfrey, Dalamitra Stella, Wiskin Louise, Taylor Martin, Univ. East Anglia, Norwich; Jones Sam, The Sheep Trust; Scarpa Riccardo, Univ. York.
Open Research
DATA AVAILABILITY STATEMENT
Dideoxy sequences of DBY, ZFY and SRY segments representing different haplogroups are accessible via the NCBI codes MF741774.1 to MF74182.1 and MG545047.1 to MG545050.1. The preliminary data set or 5356 genotypes in 70 mainly Asian and Moroccan male goats in plink format can be accessed via https://osf.io/ngx7u/?view_only=64b4f8bf78794cddb5f47a3e3e3d4534. For the VarGoats data set, see www.goatgenome.org/vargoats.html and Denoyelle et al. (2021); see PRJEB37507 for the fastq files; vcf files are available upon reasonable request.




