Phylogenomics supports incongruence between ecological specialization and taxonomy in a charismatic clade of buck moths
Abstract
Local adaptation can be a fundamental component of speciation, but its dynamics in relation to gene flow are not necessarily straightforward. Herbivorous taxa with localized host plant or habitat specialization across their geographic range are ideal models for investigating the patterns and constraints of local adaptation and its impact on diversification. The charismatic, day-flying moths of the Hemileuca maia species complex (Lepidoptera: Saturniidae) are such taxa, as they are geographically widespread, exhibit considerable ecological and morphological variability and host and habitat specificity, but apparently lack genetic differentiation across their range. Here, we use genomewide single nucleotide polymorphisms to assess relationships and population structure of this group across North America and investigate the scales where genomic divergence correlates with adaptive ecological characteristics. In contrast to previous genetic studies of the group, we find broad- and fine-scale genetic differentiation between lineages, which is at odds with various levels of taxonomic description and recognition of conservation units. Furthermore, ecological specialization only explains some fine-scale genetic differentiation, and across much of the group's range, local adaptation is apparently occurring in the face of strong gene flow. These results provide unprecedented insight into drivers of speciation in this group, the relationship between taxonomy and genomics-informed species boundaries and conservation management of internationally protected entities. Broadly, this system provides a model for understanding how local adaptation in an herbivore can arise and be maintained in the face of apparently strong gene flow, and the importance of geographic isolation in generating genomic divergence, despite a lack of ecological divergence.
1 INTRODUCTION
Patterns of fine-scale diversification across wide-ranging species or species groups can be difficult to circumscribe in biologically meaningful ways (Byun, Koop, & Reimchen, 1997; Kodandaramaiah et al., 2012; vonHoldt et al., 2011). Local adaptation should be a powerful indicator of biologically significant divergence as taxa evolve in response to relatively fine-scale ecological variables (Barrera-Moreno, Ciros-Perez, Ortega-Mayagoitia, Alcantara-Rodriguez, & Piedra-Ibarra, 2015; Hereford, 2009; Kawecki & Ebert, 2004; Tiffin & Ross-Ibarra, 2014). However, while local adaptation can be common across systems, it is often mitigated by stochastic factors (e.g., genetic drift), temporal environmental variability and gene flow (Fraser, Weir, Bernatchez, Hansen, & Taylor, 2011; Hereford, 2009; Kawecki & Ebert, 2004). Theoretically, the latter can be expected to completely inhibit local adaptation in the absence of other forms of spatially heterogenous natural selection; thus, local adaptation in systems experiencing gene flow must be the result of recent or ongoing natural selection to particular environmental factors (Kawecki & Ebert, 2004). However, it has also been demonstrated that gene flow may fail to restrict local adaptation in empirical systems, when diversifying selection is strong (e.g., Gould et al., 2014; Parchman, Buerkle, Soria-Carrasco, & Benkman, 2016; Sambatti & Rice, 2006). Thus, while local ecological specialization can lead to genetic divergence and potentially to speciation, this is not a forgone conclusion (Lenormand, 2012; Wu, 2001).
Given the complex interactions between local adaptation, gene flow and genetic differentiation, it can be instructive to critically evaluate the relationships between these characteristics in order to understand a group's diversification and the process of diversification more generally. In many groups, local morphological or ecological divergence can often be maintained without apparent corresponding genetic diversification (Barrera-Moreno et al., 2015; Joyce et al., 2009; Kodandaramaiah et al., 2012; Rubinoff, San Jose, & Peigler, 2017; Yang, Chen, Huo, & Wei, 2014), leading to questions about the mechanisms that govern local adaptation. For example, if apparent local adaptation to host or habitat does not correspond to robust genetic divergence, at what level do genotypes reflect local divergence (Gould et al., 2014; Picq, McMillan, & Puebla, 2016)? Phylogenomic and population genomic approaches have become widespread in recent years and offer powerful approaches for examining the scale at which local adaptation might operate (Tiffin & Ross-Ibarra, 2014). Understanding how patterns of genomic divergence correspond to apparent local ecological and morphological variation is crucial to understanding the broader process of diversification and the factors that constrain or promote speciation (Lenormand, 2012; Savolainen, Lascoux, & Merila, 2013; Wu, 2001). Herbivorous insects are perhaps one of the best animal models for understanding local adaptation, as they frequently exhibit clear ecological divergence in host plant use or habitat shifts, even between proximate populations exposed to the same set of ecological variables (Forister, Dyer, Singer, Stireman, & Lill, 2012; Hanks & Denno, 1994; Mopper, Beck, Simberloff, & Stiling, 1995; Thomas & Singer, 1998; Tuskes, Tuttle, & Collins, 1996).
The Hemileuca maia (Drury) species complex of moths provides an ideal model for investigating the utility of phylogenomics and population genomics to reveal fine-scale patterns of local adaptation. This group consists of large, diurnal moths that occur across eastern North America from southern Canada to Florida and west to California (Figure 1). At least nine different taxonomic names have been used to recognize species or populations in the group exhibiting varying levels of divergent morphology or host plant use (Kruse, 1998; Lemaire, 2002; Steele & Peigler, 2015; Tuskes et al., 1996), and seven entities are generally recognized currently (Rubinoff et al., 2017). Multiple populations in the northeast United States and southeast Canada are protected at both federal and state/provincial levels, including Hemileuca lucina H. Edwards and a recognized form of H. maia known as the bog buck moth, bogbean buck moth or Cryan's buck moth (Buckner, Welsh, & Sime, 2014; Environment Canada 2015; Legge, Roush, DeSalle, Vogler, & May, 1996). All members of the species group are represented by highly localized, stenotopic populations. The most widely distributed members, H. maia and Hemileuca nevadensis Stretch, feed primarily on Quercus spp. and Salicaceae, respectively, while some with narrower distributions feed on unique and diverse hosts, such as Betula spp., Ceanothus herbaceous Raf., Spirea latifolia (Ait.) Borkh. and Menyanthes trifoliata L. (Figure 1). Habitat use is also quite variable (Figure 1), ranging from bogs, wetlands, meadows and pine barrens in the Northeast and Midwest United States, to granite balds in the Appalachian Mountains, pine–oak scrub in Florida, the Loess Hills formation in Iowa, sand dunes and coastal areas of Texas, and switching to riparian areas across the American West. Although nearly all members of the group appear to need rocky or sandy soil (perhaps due to requirements of pupation, which occurs in the soil), the specific environmental conditions that limit distributions (precipitation, temperature, elevation, etc.) are unknown, and the wide distribution across the continent suggests flexibility in at least some of these parameters. The combination of habitat and host plant is commonly used to separate species that can be difficult to delimit with morphology (e.g., populations in the southwest: Peigler & Stone, 1989; Steele & Peigler, 2015). However, species limits and the identity of populations in some regions is still unclear; for example, populations in the Great Lakes region exhibit a wide range of characteristics found in H. maia, H. nevadensis, H. lucina and the bog buck moth, and most authors simply refrain from naming these populations (Figure 1; Kruse, 1998; Scholtens & Wagner, 1995, 1997). Earlier work on the species group (Legge et al., 1996; Rubinoff & Sperling, 2002, 2004; Rubinoff et al., 2017), using at most sequences of one mitochondrial and three nuclear genes, failed to find genetic divergence between a wide range of taxa that are described as distinct species or populations (Buckner et al., 2014; Kruse, 1998; Peigler & Stone, 1989; Slosser, 2001), and exhibit, in some cases, a range of obvious ecological and morphological differences maintained in near sympatry (Stamp & Bowers, 1986; Tuskes et al., 1996). Caterpillars in different populations can vary greatly not only in host use but also in levels of specialization on one or a few host plants (Gratton, 2006; Leeuw, 1974; Legge et al., 1996; Martinat, Solomon, & Leininger, 1996; Smith, 1984; Stamp & Bowers, 1986); these clear transitions in host use across geographic scales are sometimes matched by similar transitions in morphology (Scholtens & Wagner, 1997).
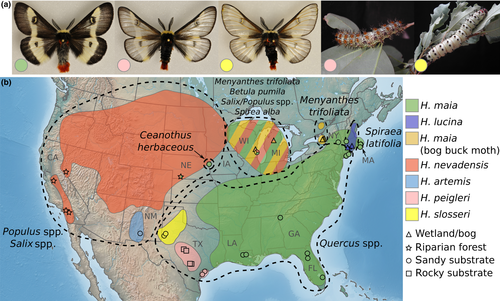
The H. maia complex presents an ideal evolutionary combination of an insect herbivore with a widespread distribution, ecological and morphological variability, and host and habitat specificity, and an apparent lack of genetic differentiation across its range (Rubinoff et al., 2017). Thus, the natural next step is to apply genomic methods to better understand the generation and maintenance of apparent local adaptation in the system. In the current study, we use genomewide single nucleotide polymorphisms (SNPs) generated with double-digest restriction-site-associated DNA sequencing (ddRAD; Peterson, Weber, Kay, Fisher, & Hoekstra, 2012) to assess population structure and genomic relationships between members of the H. maia species complex. Given the apparent lack of structure in relatively slowly evolving nuclear genes (e.g., Rubinoff et al., 2017), we ask at what scale does genomic divergence correlate with adaptive ecological characteristics for these species. To answer this question, we use a combination of population genomic and phylogenomic approaches, which can provide unprecedented resolution into the genetic structure of natural populations (Allendorf, 2017; Lemmon & Lemmon, 2013; Luikart, England, Tallmon, Jordan, & Taberlet, 2003). This integrative methodology provides a foundational framework to explore drivers of speciation in this group, the interplay between taxonomy and species boundaries, and implications of genomic data sets for management of entities of conservation concern.
2 MATERIALS AND METHODS
2.1 Specimen sampling, DNA extraction and ddRAD
Specimen sampling of adult moths and caterpillars predominately followed that of Rubinoff et al. (2017). For most specimens, we conducted new DNA extractions to provide sufficient DNA for ddRAD library preparation, and only when additional material was not available did we use prior DNA extractions. We also included new specimens to expand sampling in several geographic regions. Species determinations followed Peigler and Stone (1989) and Lemaire (2002), and detailed specimen information is provided in Supporting Information Table S1. We extracted DNA from adult thoracic tissue or cross-sectional pieces of caterpillars and homogenized tissue in tissue lysis buffer with a 3.175-mm 18/10 stainless steel bearing using a FastPrep 24 homogenizer (MP Biomedical, Santa Ana, CA) at a speed of 4.0 m/s for 20 s. Tissue homogenate was then incubated with Proteinase K following manufacturer's recommendations (Macherey-Nagel, Düren, Germany) overnight at 55°C. We finished extractions using a KingFisher Flex-96 automated extraction instrument (Thermo Scientific, Waltham, MA) with NucleoMag tissue extraction kits (Macherey-Nagel; Düren, Germany) and the optional RNase A treatment, following manufacturer's recommendations, and eluted into 100 μl of Mag-Bind elution buffer. We quantified DNA by Quant-it Picogreen assay (Thermo Fisher; Waltham, MA) on a SpectraMax M2 plate reader (Molecular Devices; Sunnyvale, CA) and used a Gilson PIPETMAX 268 (Gilson; Middleton, WI) to normalize DNA to 4 ng/μl in 44.5 μl dH2O.
We prepared ddRAD libraries following the method of Peterson et al. (2012) using ~178 ng of input DNA and restriction enzymes NlaIII and MluCI. The initial ligation was performed using 42 unique barcoded adapters, and a Blue Pippin electrophoresis unit (Sage Science; Beverly, MA) was used to size-select subpools of these 42 adapters with a 1.5% agarose gel cassette and “tight 450 bp” target size selection. We added Illumina i7 barcodes to each subpool in a final PCR and then performed a clean-up using a 1.5:1 ratio of polyethylene glycol containing solid-phase reversible immobilization beads/sample volume (DeAngelis, Wang, & Hawkins, 1995). We quantified these size-selected and cleaned products using a 2100 Bioanalyzer with the high-sensitivity DNA kit (Agilent, Santa Clara, CA) and generated a final library of 163 individuals pooled at equal molar ratios (including four blanks). This final library was sequenced using 100 bp single-end sequencing on an Illumina HiSeq 4000.
2.2 Data processing
We used the Stacks pipeline (Catchen, Amores, Hohenlohe, Cresko, & Postlethwait, 2011; Catchen, Hohenlohe, Bassham, Amores, & Cresko, 2013) to build catalog loci and call SNPs de novo. Unless specified, default settings and parameters were used for all data processing steps. First, we used process_radtags to demultiplex raw FASTQ files, discard reads with low-quality scores or uncalled bases and rescue reads that contained errors in the cut site or barcode. Then, we used denovo_map.pl to execute the main Stacks pipeline, requiring a minimum of three identical reads to create a stack and allowing for three mismatches between loci when building the catalog and when processing individuals (to accommodate higher variability between species in this data set). Finally, we used populations to generate two output formats from Stacks: vcf format containing individual SNPs for population genetic analyses and phylip format containing SNPs and flanking sequence (invariant sites of each catalog locus surrounding SNPs) for phylogenetic analyses. A minimum stack depth of eight reads per individual was required for both formats. For the vcf output, we used a population map assigning all individuals to a single population and then used vcftools version 0.1.14 (Danecek et al., 2011) for calculation of missing data per individual after excluding sites with >50% missing data. Based on this calculation of individual missingness, we then manually removed individuals with >80% missing data from the Stacks outputs and reran populations to create a new vcf file that was not biased by these poor-quality individuals and contained a single random SNP per catalog locus. We used vcftools again to create a final population genetic data file, excluding loci missing in >30% of individuals and those with a minor allele frequency (MAF) <1%. This MAF was chosen to accommodate several populations with low sample sizes (N < 6), where fixed unique alleles would have frequencies <5%. pgdspider version 2.1.0.3 (Lischer & Excoffier, 2012) was used to convert vcf format to other formats, and we refer to this data set as the “population genetic” data set. For the phylip output, we first excluded the same individuals as in the population genetic data set (identified with vcftools individual missingness calculation). We then used a population map assigning all individuals to unique populations and required loci to be present in >50% populations/individuals. We refer to this data set as the “phylogenetic” data set.
2.3 Population structure
We assessed population structure using multiple population genetic and phylogenetic approaches. First, we conducted Bayesian individual-based clustering of the population genetic data set using structure version 2.3.4 (Pritchard, Stephens, & Donnelly, 2000), which assigns individuals to genetic clusters (denoted by K) while maximizing linkage and Hardy–Weinberg equilibria. We ran 50 iterations of K = 1–20, each with 500,000 Markov chain Monte Carlo replicates (after 100,000 replicates of burn-in), and used the admixture model and correlated allele frequencies (Falush, Stephens, & Pritchard, 2003). We also conducted identical analyses using a location prior based on sampling locality, a hierarchical approach to assess substructure in the data (Vähä, Erkinaro, Niemela, & Primmer, 2007), and with the alternate ancestry prior and α = 1/10 (based on K = ~10 from preliminary analyses) as suggested by Wang (2016) for data sets with uneven sampling across populations. clumpak (Kopelman, Mayzel, Jakobsson, Rosenberg, & Mayrose, 2015) was used to average results across runs. We evaluated the optimal value of K using Ln Pr(X|K) (Pritchard et al., 2000), ΔK (Evanno, Regnaut, & Goudet, 2005) and the estimators introduced by Puechmaille (2016) (MedMedK, MedMeaK, MaxMedK and MaxMeaK, which we refer to as the “Puechmaille statistics” henceforth) implemented in structureselector (Li & Liu, 2017). For the latter, we used a population map corresponding to collection localities (generally, at the level of counties or states to avoid single individual populations) and a threshold of 0.5 (to accommodate high levels of admixed signatures found in preliminary analyses). We also used the population genetic data set to generate a NeighborNet phylogenetic network using splitstree version 4.14.6 (Huson & Bryant, 2006) with uncorrected distances.
We then used the phylogenetic data set to assess population structure using maximum-likelihood (ML) and Bayesian inference (BI) tree searches. For ML analysis, we used iq-tree version 1.6.0 (Nguyen, Schmidt, von Haeseler, & Minh, 2015), with 1,000 replicates of both ultra-fast bootstrap (Hoang, Chernomor, von Haeseler, Minh, & Vinh, 2018) and the Shimodaira–Hasegawa approximate likelihood ratio test (SH-aLRT; Guindon et al., 2010) to test node support, and used the best-fit substitution model predicted by ModelFinder (Kalyaanamoorthy, Minh, Wong, von Haeseler, & Jermiin, 2017). We used mrbayes version 3.2.6 (Huelsenbeck & Ronquist, 2001; Ronquist & Huelsenbeck, 2003) in parallel (Altekar, Dwarkadas, Huelsenbeck, & Ronquist, 2004) for BI analyses, with six runs of one million generations, each with four chains at default temperatures, sampling every 5,000 generations, and the substitution model as predicted for the ML analysis. We assessed convergence of the runs by monitoring the average standard deviation of split frequencies and the potential scale reduction factor (which should approach zero and one, respectively) and used tracer version 1.6 (Rambaut, Suchard, Xie, & Drummond, 2014) to assess effective sample sizes (which should be >200) after the analysis. We manually combined trees from independent runs, removed the first 25% of trees as burn-in and constructed a 50% majority rule consensus tree in paup version 4.0b10 (Swofford, 2002). figtree version 1.4.2 (Rambaut & Drummond, 2010) was used to visualize trees.
2.4 Descriptive statistics
We calculated general population genetic statistics (heterozygosity, inbreeding coefficients, etc.) with genodive version 2.0b27 (Meirmans & Van Tienderen, 2004), and private alleles per population with the poppr library (Kamvar, Brooks, & Grunwald, 2015; Kamvar, Tabima, & Grunwald, 2014) in r (R Core Team, 2018). We also used genodive to test for pairwise population differentiation using the molecular variance FST method (Excoffier, Smouse, & Quattro, 1992; Michalakis & Excoffier, 1996) with 10,000 permutations and to calculate pairwise Jost's D distances (Jost, 2008). Finally, we used genepop version 1.2 (Raymond & Rousset, 1995; Rousset, 2008) to calculate gene diversity among individuals in each population (1 − Q-inter). We applied Bonferroni corrections to tests with multiple comparisons.
3 RESULTS
3.1 Data properties
DNA was extracted from 137 individuals. However, initial digests produced inconsistent results for 23 individuals (confirmed by running digested products on a 1.5% agarose gel), so multiple DNA extractions and libraries were generated for these individuals, leading to a total of 163 individual libraries that were sequenced on a single lane of an Illumina HiSeq 4000. A total of 283.4 million reads were generated across all individuals, and 262.7 million of those were retained after removing reads of low-quality and ambiguous barcodes (mean per individual: 1.6 million reads). For individuals where we generated multiple DNA extractions and libraries, we selected the best library per individual by considering individual missingness calculated during the generation of the population genetic data set. This led to a total of 119 individuals being included in the population genetic and phylogenetic data sets. The population genetic data set contained 2,111 SNPs, and the phylogenetic data set contained 43,424 variable sites (11,881 Stacks catalog loci) across a 903,168-bp alignment.
3.2 Population structure
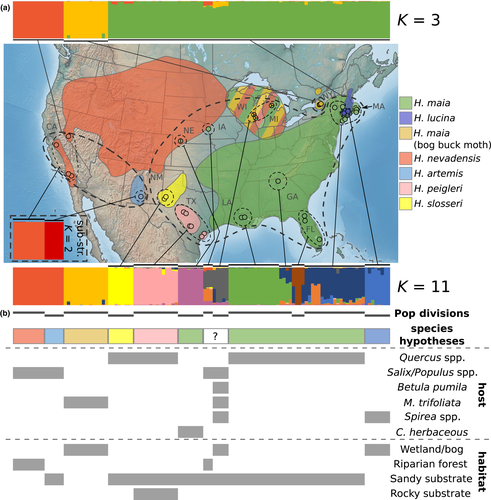
All analyses of population structure agreed on roughly the same pattern of differentiation between populations. At a broadscale, we observed high differentiation between three clusters corresponding to (a) Hemileuca artemis and Hemileuca nevadensis (b) Hemileuca maia (bog buck moth) from New York and (c) the remainder of the H. maia complex. However, at finer scales, we found support for additional clusters corresponding to the other described species and many geographically defined units (Figure 2). For the main structure analyses (all individuals, no location prior, default ancestry prior and α), ΔK supported K = 3, Ln Pr(X|K) plateaued at K = 11, and the Peuchmaille statistics generally supported K = 8 or 9. At the higher values of K, up to K = 11 where the finest-scale substructure is observed, the third aforementioned cluster is increasingly divided into distinct groups. At K = 11, Hemileuca lucina, Hemileuca peigleri and Hemileuca slosseri form distinct clusters and the remaining H. maia individuals cluster geographically: individuals from Florida, Iowa, Louisiana/Georgia, New York/Massachusetts and Wisconsin/Michigan/Nebraska form relatively distinct clusters (Figure 2). Some admixture was apparent in these groups, particularly the latter two groups, and the 11th cluster (represented by orange in the bar plot; Figure 2) was only represented at Q < 0.6. Above K = 11, no additional unique clades were observed, and the additional genetic clusters were represented by increased admixture in pre-existing clusters. structure analyses with a location prior and using the alternate ancestry prior and modified α resulted in virtually identical results as the main analysis, so we focus on the latter here. Hierarchical analysis of the main three clusters also resulted in virtually identical results as the K = 11 scenario; the only exception was the analysis of the H. artemis/H. nevadensis cluster, where K = 2 was supported, which separated the two species (Figure 2, inset). Given the overall pattern of hierarchical structure, results of other analyses (see below) and the current recommendations for structure analyses (see Janes et al., 2017 and references therein), we focus our discussion on the broadest-scale (K = 3) and finest-scale (K = 11) genetic differentiation that is supported in the data set (Figure 2). All structure results (including Ln Pr(X|K), ΔK, the Peuchmaille statistics and bar plots for each value of K) are provided in Supporting Information Figure S1.
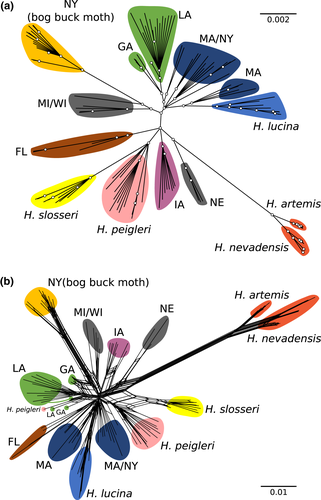
The phylogenetic network analysis and ML/BI tree searches agreed with the population structure predicted by structure, for the most part (Figure 3). The highest differentiation (both in branch length and in distance measures) was observed for clusters of H. artemis/H. nevadensis and H. maia (bog buck moth) from NY. At finer scales, most of the same clusters observed in structure were highly supported or distinct, even some of the finest-scale differentiation such as between H. artemis and H. nevadensis (Figure 3). However, some small differences were found. Populations of H. maia from MA/NY and MI/NE/WI, which were mostly contiguous clusters in the structure analyses, were split in both the phylogeny and phylogenetic network (some MA individuals were distinct from the rest of MA/NY, and MI/WI clustered apart from NE). Hemileuca peigleri was paraphyletic with regard to H. slosseri; however, the clade containing these two species was strongly supported. Finally, in the phylogeny, we observed differentiation between H. maia from GA and LA; in the phylogenetic network, we found a similar pattern but several individuals fell outside of these main groupings (including one individual of H. peigleri, although this individual did have high missing data which would bias its distance measures) (Figure 3). Consensus trees for ML and BI were virtually identical, with the only main differences being one case of more ambiguous mid-range relationships with BI (see Supporting Information Figure S2); fully labelled trees are provided in Supporting Information Figure S2.
3.3 Descriptive statistics
We used results of all of the population structure analyses to create a synthetic hypothesis of fine-scale population divisions in the group and used these divisions for calculating descriptive statistics. These predominately correspond to the K = 11 structure results; however, some clusters were further split or joined based on the phylogenetic and network analyses, leading to a total of 12 population units (Figure 2). We named these units by species when applicable and otherwise using state abbreviations for divisions within H. maia (Table 1). Heterozygosity was relatively low for all populations, ranging from 0.009 to 0.093 (Table 1), although this is largely due to the relatively low MAF; increasing the MAF to 5% resulted in measures of heterozygosity that were roughly twice as high (Supporting Information Table S2). Both inbreeding and gene diversity were variable but relatively low, ranging from 0.029 to 0.640 and 0.021 to 0.112, respectively (Table 1). Pairwise FST values were very high (average = 0.363, Table 2); however, these values were likely inflated by overall low heterozygosity and a high number of private alleles across the data set: 766 of the 4,222 (18.1%) alleles were present only in a single population (Table 1). Jost's D values are independent from heterozygosity (Jost, 2008), and exhibited less inflated values (average = 0.049), but in some cases were impacted by low sample sizes in some comparisons, leading to values of zero (Table 2). Overall, FST and Jost's D exhibited similar patterns and mimicked the K = 3 results of structure, with the highest differentiation being found in comparisons of H. artemis/H. nevadensis and New York H. maia (bog buck moth) to the remainder of the H. maia populations (Table 2).
| Population | N | H OBS | H EXP | G IS | priv | div |
|---|---|---|---|---|---|---|
| Hemileuca artemis | 6 | 0.021 | 0.022 | 0.029 | 27 | 0.021 |
| Hemileuca nevadensis | 10 | 0.009 | 0.025 | 0.640 | 52 | 0.025 |
| Hemileuca maia NY (bog buck moth) | 14 | 0.044 | 0.057 | 0.215 | 54 | 0.056 |
| Hemileuca slosseri | 8 | 0.072 | 0.085 | 0.144 | 59 | 0.083 |
| Hemileuca peigleri/maia TX | 14 | 0.093 | 0.113 | 0.174 | 115 | 0.112 |
| Hemileuca lucina | 8 | 0.077 | 0.091 | 0.157 | 32 | 0.092 |
| H. maia IA | 8 | 0.086 | 0.098 | 0.125 | 48 | 0.097 |
| H. maia FL | 4 | 0.056 | 0.090 | 0.372 | 29 | 0.090 |
| H. maia LA/GA | 20 | 0.088 | 0.103 | 0.145 | 146 | 0.102 |
| H. maia MA/NY | 19 | 0.089 | 0.112 | 0.200 | 153 | 0.111 |
| H. maia NE | 3 | 0.080 | 0.090 | 0.110 | 13 | 0.091 |
| H. maia MI/WI | 5 | 0.078 | 0.099 | 0.211 | 38 | 0.097 |
| (1) | (2) | (3) | (4) | (5) | (6) | (7) | (8) | (9) | (10) | (11) | (12) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (1) Hemileuca artemis | — | 0.000 | 0.120 | 0.089 | 0.087 | 0.097 | 0.087 | 0.000 | 0.088 | 0.082 | 0.091 | 0.091 |
| (2) Hemileuca nevadensis | 0.549 | — | 0.121 | 0.094 | 0.091 | 0.098 | 0.093 | 0.000 | 0.092 | 0.086 | 0.091 | 0.095 |
| (3) Hemileuca maia NY (bog buck moth) | 0.709 | 0.726 | — | 0.076 | 0.056 | 0.061 | 0.054 | 0.057 | 0.052 | 0.044 | 0.067 | 0.045 |
| (4) Hemileuca slosseri | 0.589 | 0.638 | 0.515 | — | 0.029 | 0.054 | 0.041 | 0.047 | 0.042 | 0.035 | 0.050 | 0.043 |
| (5) Hemileuca peigleri/maia TX | 0.484 | 0.534 | 0.384 | 0.203 | — | 0.031 | 0.020 | 0.025 | 0.021 | 0.015 | 0.031 | 0.021 |
| (6) Hemileuca lucina | 0.606 | 0.645 | 0.458 | 0.360 | 0.208 | — | 0.033 | 0.000 | 0.031 | 0.019 | 0.050 | 0.032 |
| (7) H. maia IA | 0.557 | 0.607 | 0.416 | 0.294 | 0.143 | 0.238 | — | 0.027 | 0.020 | 0.014 | 0.031 | 0.019 |
| (8) H. maia FL | 0.682 | 0.698 | 0.481 | 0.328 | 0.175 | 0.243 | 0.202 | — | 0.026 | 0.016 | 0.000 | 0.031 |
| (9) H. maia LA/GA | 0.491 | 0.534 | 0.370 | 0.284 | 0.151 | 0.217 | 0.151 | 0.187 | — | 0.011 | 0.036 | 0.023 |
| (10) H. maia MA/NY | 0.449 | 0.494 | 0.316 | 0.233 | 0.108 | 0.137 | 0.102 | 0.121 | 0.087 | — | 0.029 | 0.015 |
| (11) H. maia NE | 0.662 | 0.692 | 0.497 | 0.336 | 0.196 | 0.317 | 0.216 | 0.288 | 0.237 | 0.184 | — | 0.031 |
| (12) H. maia MI/WI | 0.603 | 0.651 | 0.393 | 0.300 | 0.149 | 0.231 | 0.146 | 0.218 | 0.169 | 0.106 | 0.212 | — |
- Bolded FST values indicate non-significant comparisons. Jost's D values of zero are a result of small sample sizes, as shown in Jost (2008).
4 DISCUSSION
Overall, our results suggest that ecological and morphological divergence in the Hemileuca maia complex are incongruous with genetic divergence. For instance, at the broadest level of population structure, that is, K = 3, populations of H. maia in northern NY, known as the bog buck moth, are differentiated from the H. maia and Hemileuca artemis/nevadensis clusters; however, virtually every non-genetic feature of the bog buck moth, including morphology, host plant use and habitat, is shared with other Great Lakes populations that, themselves, are not genetically distinct at this highest hierarchical level (Note, although Rubinoff et al. (2017) recognized Great Lakes populations of H. maia to be “bog buck moth,” henceforth, we use the name “bog buck moth” to refer only to the genomically distinct population in New York). Only when considering the finest scale of genetic differentiation are the rest of the populations that have been recognized as species across the continent considered genetically distinct (Figures 2, 3). Yet, because genetic differentiation at this finest scale also supports the distinctiveness of virtually every geographically distant population of H. maia, these results are not intuitively easy to apply since this level includes populations that exhibit little to no ecological or morphological divergence from each other. Thus, in order to recognize significant ecological specialization in host use like Ceanothus herbaceous in Iowa sand dunes and Spirea latifolia in Connecticut marshes, virtually every region across the range of H. maia must be recognized as equally distinct.
The H. maia complex exhibits clear local adaptation across its range in its host and habitat use. However, the recency and significance of many of these ecological divergences are unclear. While they may simply represent remarkable plasticity in a single, nearly transcontinental, species (Kawecki & Ebert, 2004; Sultan & Spencer, 2002), local adaptation to host plant has been demonstrated in multiple regions across this group's range which is manifested as better survivorship and growth on a population's particular host rather than alternatives used by neighbouring populations (Gratton, 2006; Legge et al., 1996; Martinat et al., 1996; Stamp & Bowers, 1986). The genetic divergence observed at the genome scale also supports local adaptation to host over phenotypic plasticity, as genetic differentiation would not be expected under adaptive phenotypic plasticity (Kawecki & Ebert, 2004), such as at the scale of H. maia across the southeast United States. Similar patterns of host plant shifts are known from other genera of moths (e.g., Hyalophora), including populations ostensibly from the same species utilizing different plant families in different mountain ranges (Tuskes et al., 1996). Alternatively, localized host plant and habitat specialization may be governed by a relatively small set of genes, which were not sampled or did not play a significant role in our phylogenomic analysis leading to a lack of reciprocal monophyly between proximate, but ecologically divergent, populations.
4.1 Drivers of speciation in the H. maia complex
While local adaptation and geographic isolation can both drive diversification, it is clear that their relative effects are remarkably heterogeneous across the members of this species complex. For instance, local adaptation to different host plant and habitat appears to be important in the isolation of Hemileuca lucina from the parapatrically distributed H. maia (Stamp & Bowers, 1986), akin to other well-known cases of selection-based ecological speciation across small geographic scales (e.g., Gasterosteus sticklebacks: Hatfield and Schluter (1999), Rhagoletis fruit flies: Filchak, Roethele, and Feder (2000) and Timema walking sticks: Nosil (2007)). The broad genetic differentiation of the H. artemis/nevadensis cluster in the West may also be explained by ecological diversification, as their exclusive feeding on Salix and Populus in riparian habitats is relatively unique. The eastern limits of this genetic entity and the extent of Salix/Populus-feeding in other regions are less clear (Scholtens & Wagner, 1995; Tietz, 1972) (discussed below).
The other instance of large genetic differentiation is less easily explained: Why does the bog buck moth exhibit such high relative genetic divergence compared to the other ecologically divergent populations? There are several potential explanations. First, the age of the lineage compared to the rest of the group could explain the higher divergence. However, bog buck moth is thought to be one of the more recent derivatives (post-glaciation) in the group (Rubinoff & Sperling, 2004; Tuskes et al., 1996), which is sensible given current understanding of the history of North American glaciation (e.g., Mickelson & Colgan, 2004); thus, greater age is unlikely to explain this divergence. Ecological divergence also fails to explain the high genetic differentiation, as there are populations in the western Great Lakes region with virtually identical morphology and ecology (e.g., Kruse, 1998), but no genetic diagnosability at the same hierarchical level. Therefore, the most likely explanation for high genetic differentiation in bog buck moth is its geographic isolation which limits gene flow. This explanation would also suggest that the other populations in the broadly distributed H. maia cluster are not physically separated to the same degree and are maintaining their ecological divergence in spite of gene flow. Taken together, our results provide a model for understanding both how local adaptation in an herbivore can arise and be maintained in the face of apparently strong gene flow and the importance of geographic isolation in generating broader genomic divergence, despite a lack of ecological divergence (Crispo, Bentzen, Reznick, Kinnison, & Hendry, 2006). While the varied nature of the diversification mechanisms shown in the H. maia species group has been seen, to a lesser extent, in other systems (e.g., Parchman et al., 2016), the complexity and variation in patterns of diversification are surprising and may be tied to evolutionarily important characteristics, such as mating systems, which are relatively indiscriminate in these moths (Collins, 2018; Peigler & Williams, 1984; Tuskes et al., 1996).
4.2 Species boundaries and taxonomy
The concept of what defines a species is fraught with inconsistencies and widely variable benchmarks (e.g., De Queiroz, 2007). Unfortunately, our results only further confirm the equivocal nature of such definitions. Specifically, the ecological divergence vs. genomic divergence contradictions suggest that there are serious inconsistencies in our concepts of species in the group, even when informed at the genomic level, to the extent that they may be counterproductive with respect to evolutionary concepts and applied uses (Drès & Mallet, 2002).
Taken at face value, the broadest scale of genetic differentiation could be used to suggest the elevation of bog buck moth in New York to species status, which would have significant management implications, since it is of conservation concern (see below). The same rationale would sink several described species, H. artemis, Hemileuca peigleri, Hemileuca slosseri and H. lucina (another species of conservation concern, see below), some of which show larger ecological and morphological divergence and distinctness than when comparing, for instance, the bog buck moth to other Menyanthes-feeding populations in the Midwestern Great Lakes region. Alternatively, at a finer genetic scale, one could argue that almost any of the small genetic clusters could be treated as distinct species or subspecies, based on varying combinations of characteristics tailored to suit each situation (e.g., genetic differentiation, host or habitat). Ultimately, even our genomics-informed view of population structure in this complex continues to highlight much of the subjectivity and debate present in the world of species/subspecies concepts (e.g., De Queiroz, 2007; Mallet, 2001; Mayden, 1997). Broadly speaking, there are obvious, important, ecologically functional differences between some populations in the H. maia group, but these differences are not broadly reflected in the genomes of the various populations and do not, apparently, inhibit ongoing gene flow between the groups. At what level such “genomic integrity” (Sperling, 2003) can be identified between genetic entities will be intrinsic to how these populations are recognized (Wu, 2001).
Despite the fact that we have sampled across the continental range of the group, there are still sampling gaps (particularly across the range of H. nevadensis s.l.: see below), and thus the potential for effects of unsampled populations on the analyses used here (“ghost populations” sensu Lawson, van Dorp, & Falush, 2018). Therefore, we are reticent to propose any of these changes formally. Rather, we hope that these results spur new interest in the relationships and population structure of the H. maia complex and geographically focused studies using genomic approaches such as that used here. Such focused treatments are particularly needed in the range of H. nevadensis. The eastern limits of this species, and its intergrade into H. maia, have remained elusive with regard to morphology, host plant and genetics (Henne & Diehl, 2002; Kruse, 1998; Legge et al., 1996; Rubinoff et al., 2017; Scholtens & Wagner, 1997; Tuskes et al., 1996). While we provide a fine-scale analysis of the species frontier in New Mexico and west Texas, where there is clear ecological and morphological distinction between H. artemis and H. slosseri, populations feeding on Salix spp. in Nebraska, which were always considered H. nevadensis based on morphology and habitat/host plant, are clearly H. maia (Figure 2). This phenomenon is repeated for populations in the Midwest where a priori identities were more ambiguous; it is clear that these populations are also H. maia. Comprehensive sampling throughout the Midwest and West will be needed to elucidate the taxonomic and evolutionary situation in this unsampled region.
4.3 Management implications
Two taxa in this group, H. lucina and the bog buck moth, are of conservation concern in the United States, and for the latter, Canada (Buckner et al., 2014; Environment Canada 2015; Legge et al., 1996; Rubinoff & Sperling, 2004), thus giving these results, and their interpretation, applied significance. While H. lucina is described as a distinct species and has clear ecological differences (host plant in particular), it is only genetically distinct at the finest scale of differentiation (Figures 2, 3). If we consider this level of genetic differentiation, then virtually every geographic cluster of populations in the species complex becomes equally distinct taxa. Thus, from a systematic perspective, we either disregard the ecological uniqueness of H. lucina or we recognize that every cluster of populations in the complex that we sampled represents a distinct entity, even those across the eastern third of the continent that exhibit very limited to no morphological and ecological differentiation. The bog buck moth complicates this decision because, while not yet described as a species, it is distinct at the broadest scale of genetic differentiation considered here, reflecting much deeper divergence than H. lucina. This deep divergence may be explained by the isolation of bog buck moth to the north of most H. maia populations, vs. the potential for genetic exchange between H. lucina and H. maia given their parapatric distribution throughout the range of H. lucina. As discussed above, this suggests that the genetic underpinnings for H. lucina's ecological specialization are maintained in the face of strong gene flow from H. maia, which has obscured the distinct nature of the former at all but the finest levels of genetic differentiation.
This discrepancy between taxonomic status and genetic divergence of H. lucina and bog buck moth presents a particular interesting implication for federal conservation protection, as invertebrates must be described subspecies or species to be federally protected in the United States (Waples, Nammack, Cochrane, & Hutchings, 2013). Thus, H. lucina would currently be eligible for federal protection, while bog buck moth would not, although state protections do not generally have the same limitation. Management of population units below the species level is generally accomplished through identification of evolutionarily significant units, which are populations exhibiting genetic differentiation and adaptive variability that would warrant protection (Crandall, Bininda-Emonds, Mace, & Wayne, 2000; Fraser & Bernatchez, 2001; Pearse, 2016; Shafer et al., 2015). The presented genomic results definitely satisfy the criteria of genetic differentiation for bog buck moth, and the use of Menyanthes trifoliata may satisfy that of adaptive variability. However, the latter is not exclusive to the genetic cluster identified as bog buck moth, as populations in Michigan (Scholtens & Wagner, 1997) and Wisconsin (Kruse, 1998) also feed on M. trifoliata. More comprehensive sampling of bog buck moth (particularly Canadian populations), and more populations in Michigan and Wisconsin, will be important to build on these results and our knowledge of the extent of broad- and fine-scale genetic differentiation between these populations.
5 CONCLUSIONS
Here, we present the most widely sampled genetic assessment of the Hemileuca maia complex to date. Contrary to the previous genetic studies, our genomewide SNP data set supports high levels of hierarchical differentiation across North America. This hierarchical structure is inconsistent with the current taxonomy of the group and is only partially concordant with ecological specialization, suggesting that ecological adaptation is occurring in the face of strong gene flow. In the case of bog buck moth, a population of conservation concern, geographic isolation has seemingly led to genomic divergence without complete ecological differentiation. Thus, this system provides a model for investigating the effects of both local ecological adaptation and geographic isolation without ecological divergence on the process of diversification. Further studies, guided by these results and using more localized, larger and finer-scale sampling regimes, will be required to dig deeper into these evolutionarily complex cases of local adaptation.
ACKNOWLEDGEMENTS
Library preparation was performed at the United States Department of Agriculture—Agricultural Research Service (USDA-ARS) Daniel K. Inouye US PBARC Genomics facility by Nicole Yoneishi, and sequencing was conducted at the Vincent J. Coates Genomics Sequencing Laboratory at University of California at Berkeley, supported by National Institutes of Health S10 Instrumentation Grants S10RR029668 and S10RR027303. We thank J. Adams, A. Brees, C. Buelow, D. Bustos, A. Cognato, M. Denoux, P. Goldstein, A. Hammond, S. Johnson, J. Kruse, R. Lyttle, T. McCabe, S. McElfresh, E. Metzler, R. Nuelle, K. Osborne, M. Smith, S. Spomer, T. Steele, E. Stanton, D. Wagner and A. Warren for assistance in collecting specimens. Partial funding was provided by USDA-ARS, the College of Tropical Agriculture and Human Resources, University of Hawaii at Manoa, and USDA Cooperative State Research, Education and Extension (CSREES) project HAW00942-H administered by the College of Tropical Agriculture and Human Resources, University of Hawaii. USDA is an equal opportunity employer. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA.
DATA ACCESSIBILITY
Demultiplexed data files are available as a Sequence Read Archive (National Center for Biotechnology Information) under accession SRP127729 (BioProject PRJNA427837, SRA SRR6429876–SRR6430038), and analysis input files and supplementary files are available at Dryad accession https://doi.org/10.5061/dryad.q4p27mb.
AUTHOR CONTRIBUTION
All authors conceptualized the study. R.S.P. and D.R. gathered the specimens, J.R.D. and S.M.G. generated genomic data, and J.R.D. conducted analyses. J.R.D. and D.R. wrote the manuscript with input from all authors.




